Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Trophoblast Sample Collection
2.2. Hematoxylin-Eosin and Immunohistochemistry Staining
2.3. RNA Extraction and Quality Controls
2.4. Library Preparation for Transcriptome Sequencing and Bioinformatics Analysis
2.5. Gene Expression Level Quantification, Commonly Expressed Genes (CEGs), and Differently Expressed Genes (DEGs) Analysis
2.6. GO, KEGG, and PPI Enrichment Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR) for RNA Sequence Validation
2.8. Protein Extraction and Western Blotting
2.9. Statistical Analysis
3. Results
3.1. Hircine Trophoblast Showed Significant Growth, Angiogenesis, and Adhesion at Early Pregnancy Stages
3.2. Identification of Expressed Transcripts in the Hircine Trophoblast Membrane Transcriptome
3.3. The Top 100 CEGs of Trophoblast Were Highly Related to Protein Translation and Placental Development
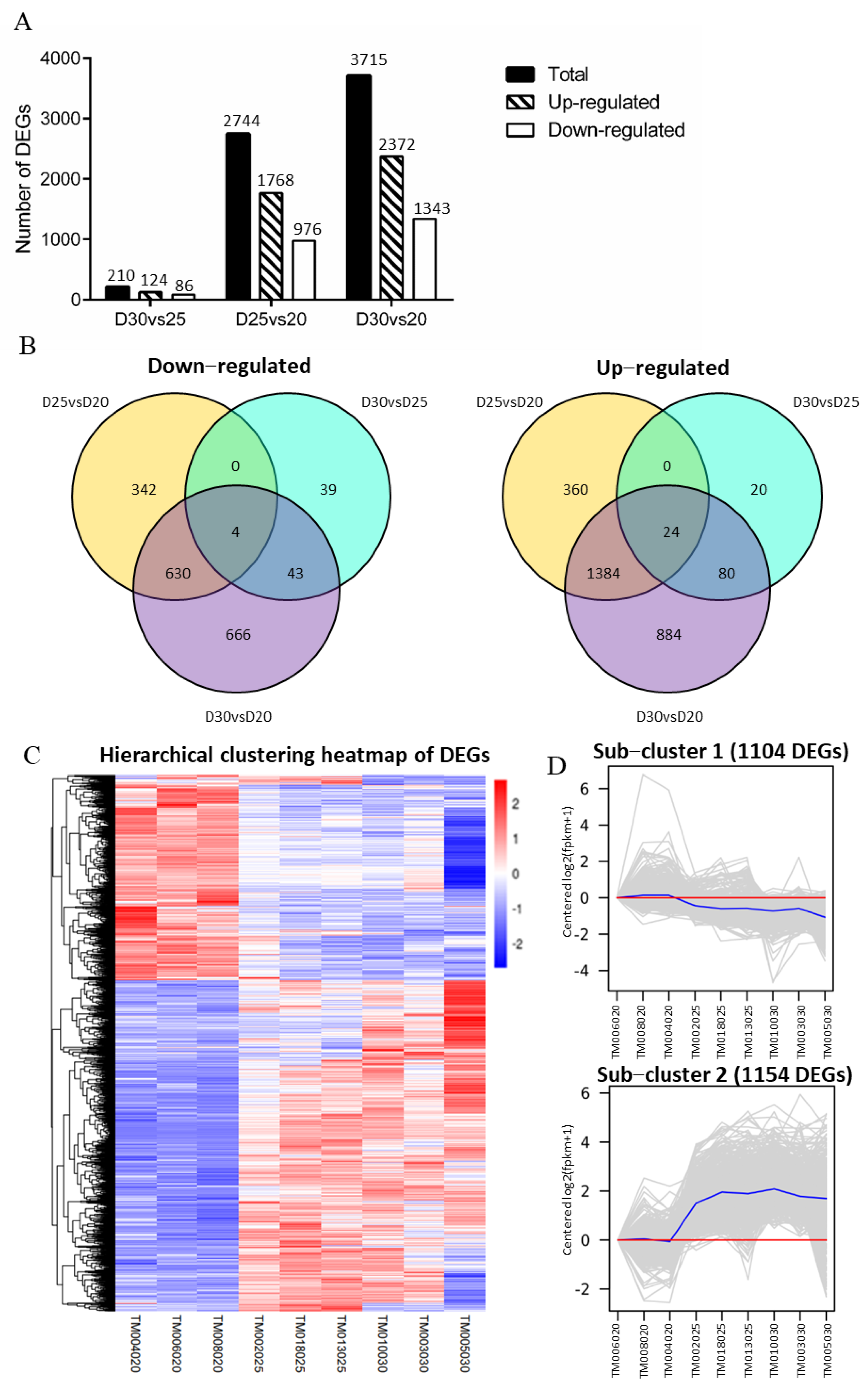
3.4. Identifying DEGs on Trophoblast Membranes among the Three-Time Points
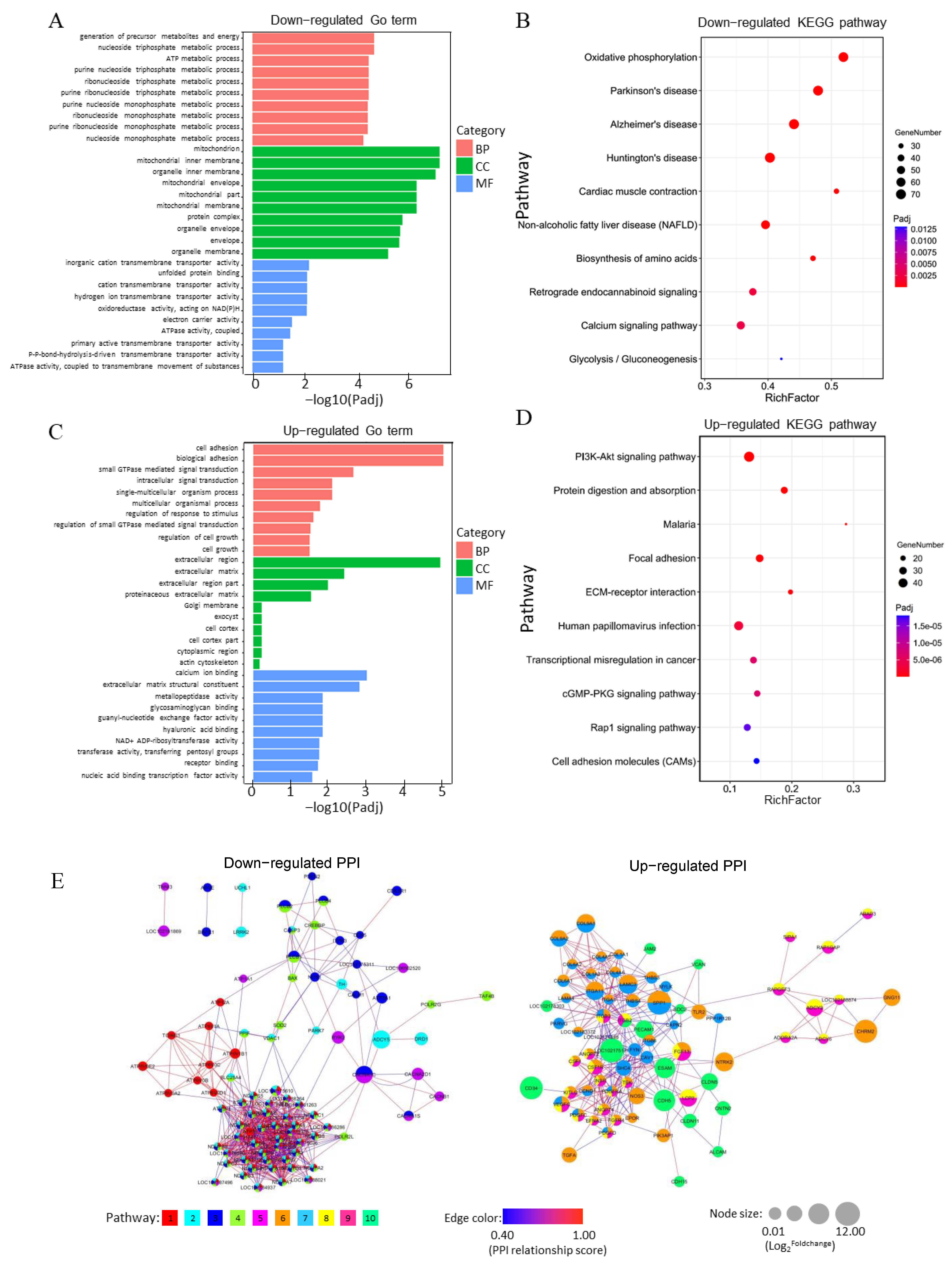
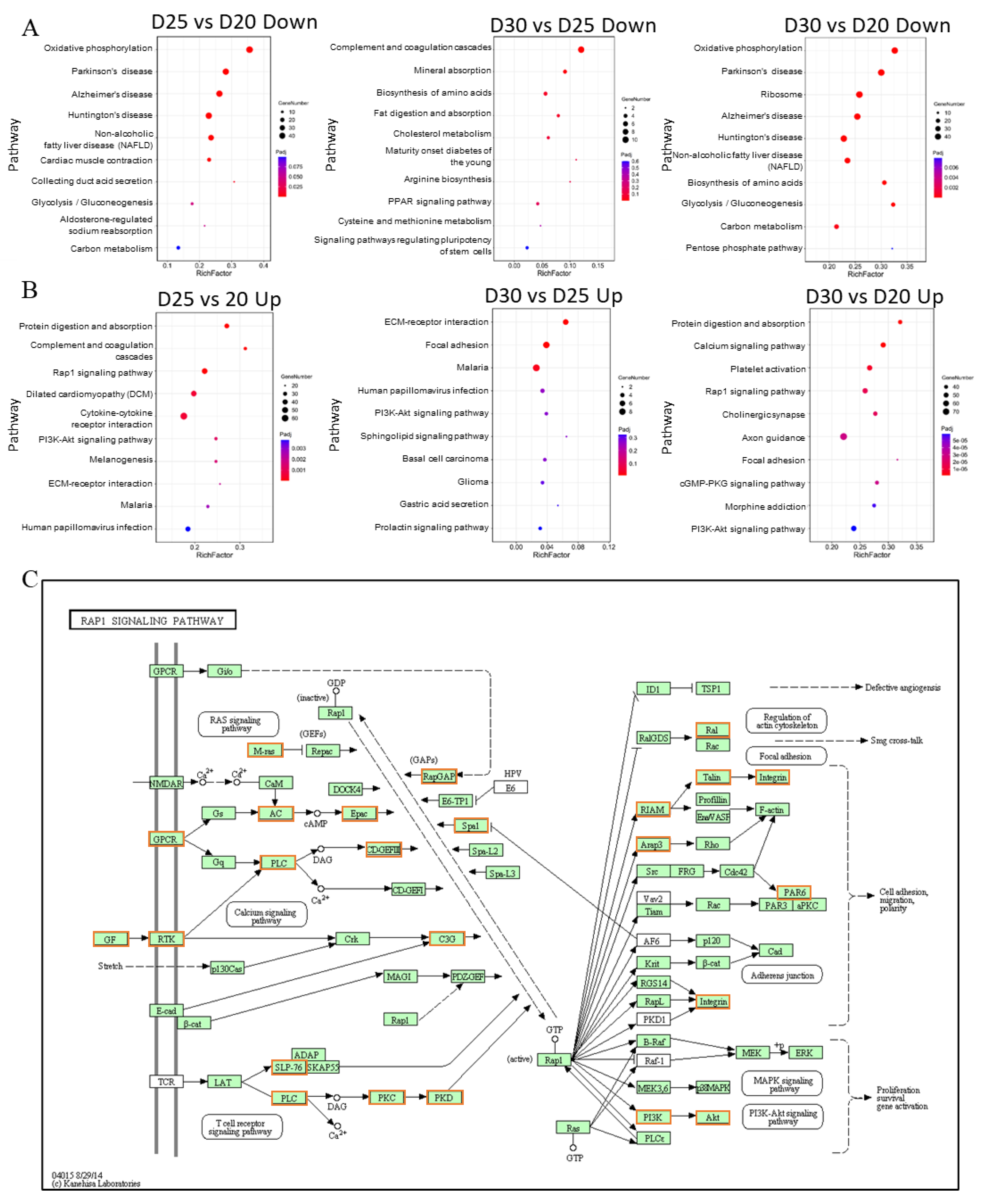
3.5. Analysis of Functional Annotation and Pathway Enrichment in the Main Clusters of DEGs
3.6. Rap1 Signaling Pathway Connects with Other Functional Pathways to Regulate Placental Development
3.7. Validation of DEGs Expression and Summary of Physiological Processes in the Trophoblast Membrane of Goats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Burton, G.J.; Fowden, A.L. The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef]
- Salomon, C.; Rice, G.E. Role of Exosomes in Placental Homeostasis and Pregnancy Disorders. Prog. Mol. Biol. Transl. Sci. 2017, 145, 163–179. [Google Scholar] [CrossRef]
- Angiolini, E.; Fowden, A.; Coan, P.; Sandovici, I.; Smith, P.; Dean, W.; Burton, G.; Tycko, B.; Reik, W.; Sibley, C.; et al. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta 2006, 27, S98–S102. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.J.; Sheehan, P.M.; Brennecke, S.P.; Keogh, R.J. Vessel remodelling, pregnancy hormones and extravillous trophoblast function. Mol. Cell. Endocrinol. 2012, 349, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Colucci, F.; Bouenouar, S.; Kieckbusch, J.; Moffett, A. How does variability of immune system genes affect placentation? Placenta 2011, 32, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Fisher, S.J. Trophoblast stem cells. Biol. Reprod. 2011, 84, 412–421. [Google Scholar] [CrossRef]
- Knofler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.; James, J. Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef]
- Reynolds, L.P.; Caton, J.S.; Redmer, D.A.; Grazul-Bilska, A.T.; Vonnahme, K.A.; Borowicz, P.P.; Luther, J.S.; Wallace, J.M.; Wu, G.; Spencer, T.E. Evidence for altered placental blood flow and vascularity in compromised pregnancies. J. Physiol. 2006, 572, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, L.P.; Borowicz, P.P.; Caton, J.S.; Vonnahme, K.A.; Luther, J.S.; Buchanan, D.S.; Hafez, S.A.; Grazul-Bilska, A.T.; Redmer, D.A. Uteroplacental vascular development and placental function: An update. Int. J. Dev. Biol. 2010, 54, 355–365. [Google Scholar] [CrossRef]
- Chang, T.A.; Bondarenko, G.I.; Gerami-Naini, B.; Drenzek, J.G.; Durning, M.; Garthwaite, M.A.; Schmidt, J.K.; Golos, T.G. Trophoblast differentiation, invasion and hormone secretion in a three-dimensional in vitro implantation model with rhesus monkey embryos. Reprod. Biol. Endocrinol. 2018, 16, 24. [Google Scholar] [CrossRef]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W. Osteopontin: A leading candidate adhesion molecule for implantation in pigs and sheep. J. Anim. Sci. Biotechnol. 2014, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Wakeland, A.K.; Parast, M.M. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J. Endocrinol. 2018, 236, R43–R56. [Google Scholar] [CrossRef] [PubMed]
- West, R.C.; Ming, H.; Logsdon, D.M.; Sun, J.; Rajput, S.K.; Kile, R.A.; Schoolcraft, W.B.; Roberts, R.M.; Krisher, R.L.; Jiang, Z.; et al. Dynamics of trophoblast differentiation in peri-implantation-stage human embryos. Proc. Natl. Acad. Sci. USA 2019, 116, 22635–22644. [Google Scholar] [CrossRef]
- Charnock-Jones, D.S.; Kaufmann, P.; Mayhew, T.M. Aspects of human fetoplacental vasculogenesis and angiogenesis. I. Molecular regulation. Placenta 2004, 25, 103–113. [Google Scholar] [CrossRef]
- Chen, D.B.; Feng, L.; Hodges, J.K.; Lechuga, T.J.; Zhang, H. Human trophoblast-derived hydrogen sulfide stimulates placental artery endothelial cell angiogenesis. Biol. Reprod. 2017, 97, 478–489. [Google Scholar] [CrossRef]
- O’Connell, A.R.; Demmers, K.J.; Smaill, B.; Reader, K.L.; Juengel, J.L. Early embryo loss, morphology, and effect of previous immunization against androstenedione in the ewe. Theriogenology 2016, 86, 1285–1293. [Google Scholar] [CrossRef]
- Dunne, L.D.; Diskin, M.G.; Sreenan, J.M. Embryo and foetal loss in beef heifers between day 14 of gestation and full term. Anim. Reprod. Sci. 2000, 58, 39–44. [Google Scholar] [CrossRef]
- Luo, N.J.; Wang, J.; Hu, Y.; Zhao, Z.Q.; Zhao, Y.J.; Chen, X.C. Cold and heat climatic variations reduce indigenous goat birth weight and enhance pre-weaning mortality in subtropical monsoon region of China. Trop. Anim. Health Prod. 2020, 52, 1385–1394. [Google Scholar] [CrossRef]
- Han, Y.G.; Zeng, Y.; Huang, Y.F.; Huang, D.L.; Peng, P.; Na, R.S. A nonsynonymous SNP within the AMH gene is associated with litter size in Dazu black goats. Anim. Biotechnol. 2020, 5, 1–5. [Google Scholar] [CrossRef]
- Guillomot, M. Cellular Interactions during Implantation in Domestic Ruminants. J. Reprod. Fertil. 1995, 49, 39–51. [Google Scholar] [CrossRef]
- Spencer, T.E.; Johnson, G.A.; Bazer, F.W.; Burghardt, R.C. Implantation mechanisms: Insights from the sheep. Reproduction 2004, 128, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Guillomot, M.; Flechon, J.E.; Wintenbergertorres, S. Conceptus Attachment in the Ewe—An Ultrastructural-Study. Placenta 1981, 2, 169–181. [Google Scholar] [CrossRef]
- Grazul-Bilska, A.T.; Borowicz, P.P.; Johnson, M.L.; Minten, M.A.; Bilski, J.J.; Wroblewski, R.; Redmer, D.A.; Reynolds, L.P. Placental development during early pregnancy in sheep: Vascular growth and expression of angiogenic factors in maternal placenta. Reproduction 2010, 140, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Grazul-Bilska, A.T.; Johnson, M.L.; Borowicz, P.P.; Minten, M.; Bilski, J.J.; Wroblewski, R.; Velimirovich, M.; Coupe, L.R.; Redmer, D.A.; Reynolds, L.P. Placental development during early pregnancy in sheep: Cell proliferation, global methylation, and angiogenesis in the fetal placenta. Reproduction 2011, 141, 529–540. [Google Scholar] [CrossRef]
- Lucy, M.C.; Evans, T.J.; Poock, S.E. Lymphocytic foci in the endometrium of pregnant dairy cows: Characterization and association with reduced placental weight and embryonic loss. Theriogenology 2016, 86, 1711–1719. [Google Scholar] [CrossRef]
- Blomberg, L.; Hashizume, K.; Viebahn, C. Blastocyst elongation, trophoblastic differentiation, and embryonic pattern formation. Reproduction 2008, 135, 181–195. [Google Scholar] [CrossRef]
- Horcajo, P.; Jimenez-Pelayo, L.; Garcia-Sanchez, M.; Regidor-Cerrillo, J.; Collantes-Fernandez, E.; Rozas, D.; Hambruch, N.; Pfarrer, C.; Ortega-Mora, L.M. Transcriptome modulation of bovine trophoblast cells in vitro by Neospora caninum. Int. J. Parasitol. 2017, 47, 791–799. [Google Scholar] [CrossRef]
- Wei, X.; Xiaoling, Z.; Kai, M.; Rui, W.; Jing, X.; Min, G.; Zhonghong, W.; Jianhui, T.; Xinyu, Z.; Lei, A. Characterization and comparative analyses of transcriptomes for in vivo and in vitro produced peri-implantation conceptuses and endometria from sheep. J. Reprod. Dev. 2016, 62, 279–287. [Google Scholar] [CrossRef]
- Rosario, F.J.; Powell, T.L.; Gupta, M.B.; Cox, L.; Jansson, T. mTORC1 Transcriptional Regulation of Ribosome Subunits, Protein Synthesis, and Molecular Transport in Primary Human Trophoblast Cells. Front. Cell. Dev. Biol. 2020, 8, 583801. [Google Scholar] [CrossRef] [PubMed]
- Arai, K.Y.; Nishiyama, T. Developmental changes in extracellular matrix messenger RNAs in the mouse placenta during the second half of pregnancy: Possible factors involved in the regulation of placental extracellular matrix expression. Biol. Reprod. 2007, 77, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Nasu, K.; Fujisawa, K.; Nishida, Y.; Kai, S.; Sugano, T.; Miyakawa, I.; Tateishi, Y. Expression of collagen XVIII mRNA and protein in human umbilical vein and placenta. Reprod. Fertil. Dev. 2003, 15, 107–114. [Google Scholar] [CrossRef]
- Song, G.; Bailey, D.W.; Dunlap, K.A.; Burghardt, R.C.; Spencer, T.E.; Bazer, F.W.; Johnson, G.A. Cathepsin B, Cathepsin L, and Cystatin C in the Porcine Uterus and Placenta: Potential Roles in Endometrial/Placental Remodeling and in Fluid-Phase Transport of Proteins Secreted by Uterine Epithelia Across Placental Areolae. Biol. Reprod. 2010, 82, 854–864. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, G.; Li, J.X. Effect of hypoxia on expression of placental trophoblast cells SATB1 and beta-catenin and its correlation with the pathogenesis of preeclampsia. Asian Pac. J. Trop. Med. 2016, 9, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Tandiya, U.; Nagar, V.; Yadav, V.P.; Ali, I.; Gupta, M.; Dangi, S.S.; Hyder, I.; Yadav, B.; Bhakat, M.; Chouhan, V.S.; et al. Temporal changes in pregnancy-associated glycoproteins across different stages of gestation in the Barbari goat. Anim. Reprod. Sci. 2013, 142, 141–148. [Google Scholar] [CrossRef]
- Melhem, H.; Kallol, S.; Huang, X.; Luthi, M.; Ontsouka, C.E.; Keogh, A.; Stroka, D.; Thormann, W.; Schneider, H.; Albrecht, C. Placental secretion of apolipoprotein A1 and E: The anti-atherogenic impact of the placenta. Sci. Rep. 2019, 9, 6225. [Google Scholar] [CrossRef]
- Arcuri, F.; Papa, S.; Meini, A.; Carducci, A.; Romagnoli, R.; Bianchi, L.; Riparbelli, M.G.; Sanchez, J.C.; Palmi, M.; Tosi, P.; et al. The translationally controlled tumor protein is a novel calcium binding protein of the human placenta and regulates calcium handling in trophoblast cells. Biol. Reprod. 2005, 73, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Ikezaki, M.; Toujima, S.; Iwahashi, N.; Mizoguchi, M.; Nanjo, S.; Minami, S.; Ihara, Y.; Ino, K. Calreticulin Is Involved in Invasion of Human Extravillous Trophoblasts Through Functional Regulation of Integrin beta1. Endocrinology 2017, 158, 3874–3889. [Google Scholar] [CrossRef] [PubMed]
- Iwahashi, N.; Ikezaki, M.; Matsuzaki, I.; Yamamoto, M.; Toujima, S.; Murata, S.I.; Ihara, Y.; Ino, K. Calreticulin Regulates Syncytialization Through Control of the Synthesis and Transportation of E-Cadherin in BeWo Cells. Endocrinology 2019, 160, 359–374. [Google Scholar] [CrossRef]
- MacLean, J.A., 2nd; Chakrabarty, A.; Xie, S.; Bixby, J.A.; Roberts, R.M.; Green, J.A. Family of Kunitz proteins from trophoblast: Expression of the trophoblast Kunitz domain proteins (TKDP) in cattle and sheep. Mol. Reprod. Dev. 2003, 65, 30–40. [Google Scholar] [CrossRef]
- Chakrabarty, A.; Roberts, M.R. Ets-2 and C/EBP-beta are important mediators of ovine trophoblast Kunitz domain protein-1 gene expression in trophoblast. BMC Mol. Biol. 2007, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Hong, L.; Liu, R.; Chen, R.; Li, X.; Yu, M. Cellular Localization and Regulation of Expression of the PLET1 Gene in Porcine Placenta. Int. J. Mol. Sci. 2016, 17, 2048. [Google Scholar] [CrossRef]
- Awad, M.; Koshi, K.; Kizaki, K.; Takahashi, T.; Hashizume, K. SOLD1 is expressed in bovine trophoblast cell lines and regulates cell invasiveness. Reprod. Biol. Endocrinol. 2014, 12, 55. [Google Scholar] [CrossRef]
- Ushizawa, K.; Takahashi, T.; Hosoe, M.; Kizaki, K.; Hashizume, K. Cloning and expression of SOLD1 in ovine and caprine placenta, and their expected roles during the development of placentomes. BMC Dev. Biol. 2010, 10, 9. [Google Scholar] [CrossRef]
- Ushizawa, K.; Takahashi, T.; Hosoe, M.; Kizaki, K.; Hashizume, K. Characterization and Expression Analysis of SOLD1, a Novel Member of the Retrotransposon-Derived Ly-6 Superfamily, in Bovine Placental Villi. PLoS ONE 2009, 4, e5814. [Google Scholar] [CrossRef]
- Wang, C.C.; Lo, H.F.; Lin, S.Y.; Chen, H. RACK1 (receptor for activated C-kinase 1) interacts with FBW2 (F-box and WD-repeat domain-containing 2) to up-regulate GCM1 (glial cell missing 1) stability and placental cell migration and invasion. Biochem. J. 2013, 453, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Gomez, T.; Quiñonero, A.; Dominguez, F.; Rubert, L.; Perales, A.; Hajjar, K.A.; Simon, C. Preeclampsia: A defect in decidualization is associated with deficiency of Annexin A2. Am. J. Obstet. Gynecol. 2020, 222, e1–e376. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Nowak, R.A. The role of basigin in reproduction. Reproduction 2019, 159, R97–R109. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.J.; Diao, H.L.; Ma, X.H.; Ding, N.Z.; Kadomatsu, K.; Muramatsu, T.; Yang, Z.M. Basigin expression and hormonal regulation in the rat uterus during the peri-implantation period. Reproduction 2002, 124, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Lievano, S.; Alarcon, L.; Chavez-Munguia, B.; Gonzalez-Mariscal, L. Endothelia of term human placentae display diminished expression of tight junction proteins during preeclampsia. Cell. Tissue Res. 2006, 324, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Blair, J.D.; Yuen, R.K.; Robinson, W.P.; von Dadelszen, P. Genome-wide DNA methylation identifies trophoblast invasion-related genes: Claudin-4 and Fucosyltransferase IV control mobility via altering matrix metalloproteinase activity. Mol. Hum. Reprod. 2015, 21, 452–465. [Google Scholar] [CrossRef]
- Wang, H.X.; Wang, H.M.; Lin, H.Y.; Yang, Q.; Zhang, H.; Tsang, B.K.; Zhu, C. Proteasome subunit LMP2 is required for matrix metalloproteinase-2 and -9 expression and activities in human invasive extravillous trophoblast cell line. J. Cell. Physiol. 2006, 206, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Vanderpuye, O.A.; Labarrere, C.A.; McIntyre, J.A. Predominant expression of the beta subunit of prolyl 4-hydroxylase (disulfide isomerase) in human extravillous trophoblasts. Histochemistry 1993, 100, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.H.; Wei, J.; Lu, M.Q.; Jin, M.Y.; Geng, H.L. Protective effect of human umbilical cord mesenchymal stem cell exosomes on preserving the morphology and angiogenesis of placenta in rats with preeclampsia. Biomed. Pharm. 2018, 105, 1240–1247. [Google Scholar] [CrossRef]
- You, J.L.; Wang, W.; Tang, M.Y.; Ye, Y.H.; Liu, A.X.; Zhu, Y.M. A potential role of galectin-1 in promoting mouse trophoblast stem cell differentiation. Mol. Cell. Endocrinol. 2018, 470, 228–239. [Google Scholar] [CrossRef]
- Toudic, C.; Vargas, A.; Xiao, Y.; St-Pierre, G.; Bannert, N.; Lafond, J.; Rassart, E.; Sato, S.; Barbeau, B. Galectin-1 interacts with the human endogenous retroviral envelope protein syncytin-2 and potentiates trophoblast fusion in humans. FASEB J. 2019, 33, 12873–12887. [Google Scholar] [CrossRef]
- Johnson, G.A.; Burghardt, R.C.; Bazer, F.W.; Spencer, T.E. Osteopontin: Roles in implantation and placentation. Biol. Reprod. 2003, 69, 1458–1471. [Google Scholar] [CrossRef]
- Chiok, K.L.R.; Shah, D.H. Identification of common highly expressed genes of Salmonella Enteritidis by in silico prediction of gene expression and in vitro transcriptomic analysis. Poult. Sci. 2019, 98, 2948–2963. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.H.; Lee, Y.S.; Hwang, S.M. Transcriptome analysis of common gene expression in human mesenchymal stem cells derived from four different origins. Methods. Mol. Biol. 2011, 698, 405–417. [Google Scholar] [CrossRef]
- Kang, X.; Liu, G.; Liu, Y.; Xu, Q.; Zhang, M.; Fang, M. Transcriptome profile at different physiological stages reveals potential mode for curly fleece in Chinese tan sheep. PLoS ONE 2013, 8, e71763. [Google Scholar] [CrossRef]
- Patel, J.; Landers, K.; Mortimer, R.H.; Richard, K. Regulation of Hypoxia Inducible Factors (HIF) in Hypoxia and Normoxia During Placental Development. Placenta 2010, 31, 951–957. [Google Scholar] [CrossRef]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell. Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef]
- Anton, L.; DeVine, A.; Polyak, E.; Olarerin-George, A.; Brown, A.G.; Falk, M.J.; Elovitz, M.A. HIF-1alpha Stabilization Increases miR-210 Eliciting First Trimester Extravillous Trophoblast Mitochondrial Dysfunction. Front. Physiol. 2019, 10, 699. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Yu, Q.; Jia, B.; Zhou, W.; Zhang, Y.; Mu, L. HIF3alpha affects preeclampsia development by regulating EVT growth via activation of the Flt1/JAK/STAT signaling pathway in hypoxia. Mol. Med. Rep. 2021, 23, 68. [Google Scholar] [CrossRef] [PubMed]
- Wakeland, A.K.; Soncin, F.; Moretto-Zita, M.; Chang, C.W.; Horii, M.; Pizzo, D.; Nelson, K.K.; Laurent, L.C.; Parast, M.M. Hypoxia Directs Human Extravillous Trophoblast Differentiation in a Hypoxia-Inducible Factor-Dependent Manner. Am. J. Pathol. 2017, 187, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Zimna, A.; Kurpisz, M. Hypoxia-Inducible Factor-1 in Physiological and Pathophysiological Angiogenesis: Applications and Therapies. Biomed. Res. Int. 2015, 2015, 549412. [Google Scholar] [CrossRef]
- Richani, K.; Soto, E.; Romero, R.; Espinoza, J.; Chaiworapongsa, T.; Nien, J.K.; Edwin, S.; Kim, Y.M.; Hong, J.S.; Mazor, M. Normal pregnancy is characterized by systemic activation of the complement system. J. Matern. Fetal Neonatal Med. 2005, 17, 239–245. [Google Scholar] [CrossRef]
- Mitchell, M.D.; Peiris, H.N.; Kobayashi, M.; Koh, Y.Q.; Duncombe, G.; Illanes, S.E.; Rice, G.E.; Salomon, C. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015, 213, S173–S181. [Google Scholar] [CrossRef]
- Lynch, A.M.; Murphy, J.R.; Byers, T.; Gibbs, R.S.; Neville, M.C.; Giclas, P.C.; Salmon, J.E.; Holers, V.M. Alternative complement pathway activation fragment Bb in early pregnancy as a predictor of preeclampsia. Am. J. Obstet. Gynecol. 2008, 198, 385.e1–385.e9. [Google Scholar] [CrossRef] [PubMed]
- Lynch, A.M.; Gibbs, R.S.; Murphy, J.R.; Giclas, P.C.; Salmon, J.E.; Holers, V.M. Early Elevations of the Complement Activation Fragment C3a and Adverse Pregnancy Outcomes. Obstet. Gynecol. 2011, 117, 75–83. [Google Scholar] [CrossRef]
- Chatzizacharias, N.A.; Kouraklis, G.P.; Theocharis, S.E. The role of focal adhesion kinase in early development. Histol. Histopathol. 2010, 25, 1039–1055. [Google Scholar] [CrossRef] [PubMed]
- Tapial Martinez, P.; Lopez Navajas, P.; Lietha, D. FAK Structure and Regulation by Membrane Interactions and Force in Focal Adhesions. Biomolecules 2020, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.; Artym, V.V.; Green, J.A.; Yamada, K.M. The matrix reorganized: Extracellular matrix remodeling and integrin signaling. Curr. Opin. Cell. Biol. 2006, 18, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Frank, J.W.; Burghardt, R.C.; Bazer, F.W.; Johnson, G.A. Integrins and OPN localize to adhesion complexes during placentation in sheep. Reproduction 2020, 160, 521–532. [Google Scholar] [CrossRef]
- Burghardt, R.C.; Burghardt, J.R.; Taylor, J.D.; Reeder, A.T.; Nguen, B.T.; Spencer, T.E.; Bayless, K.J.; Johnson, G.A. Enhanced focal adhesion assembly reflects increased mechanosensation and mechanotransduction at maternal-conceptus interface and uterine wall during ovine pregnancy. Reproduction 2009, 137, 567–582. [Google Scholar] [CrossRef]
- Liu, C.; Liang, X.; Wang, J.; Zheng, Q.; Zhao, Y.; Khan, M.N.; Liu, S.; Yan, Q. Protein O-fucosyltransferase 1 promotes trophoblast cell proliferation through activation of MAPK and PI3K/Akt signaling pathways. Biomed. Pharm. 2017, 88, 95–101. [Google Scholar] [CrossRef]
- Xu, Y.L.; Sui, L.L.; Qiu, B.T.; Yin, X.J.; Liu, J.T.; Zhang, X.H. ANXA4 promotes trophoblast invasion via the PI3K/Akt/eNOS pathway in preeclampsia. Am. J. Physiol. Cell. Physiol. 2019, 316, C481–C491. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Zhang, Y.; Qu, H.M.; Xu, F.S.; Hu, H.Y.; Zhang, Q.; Ye, Y.H. Reduced ELABELA expression attenuates trophoblast invasion through the PI3K/AKT/mTOR pathway in early onset preeclampsia. Placenta 2019, 87, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.M.; Zhang, Z.Z.; Ma, X.; Fang, S.F.; Qin, X.H. Repression of microRNA-21 inhibits retinal vascular endothelial cell growth and angiogenesis via PTEN dependent-PI3K/Akt/VEGF signaling pathway in diabetic retinopathy. Exp. Eye. Res. 2020, 190, 107886. [Google Scholar] [CrossRef]
- Shah, S.; Brock, E.J.; Ji, K.; Mattingly, R.R. Ras and Rap1: A tale of two GTPases. Semin. Cancer Biol. 2019, 54, 29–39. [Google Scholar] [CrossRef]
- Kusama, K.; Yoshie, M.; Tamura, K.; Daikoku, T.; Takarada, T.; Tachikawa, E. Possible roles of the cAMP-mediators EPAC and RAP1 in decidualization of rat uterus. Reproduction 2014, 147, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Huang, J.P.; Chu, T.Y.; Aplin, J.D.; Chen, C.Y.; Wu, Y.H. Human placental multipotent mesenchymal stromal cells modulate trophoblast migration via Rap1 activation. Placenta 2013, 34, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Cheong, M.L.; Chang, G.D.; Tsai, M.S.; Chen, H.W. Involvement of Epac1/Rap1/CaMKI/HDAC5 signaling cascade in the regulation of placental cell fusion. Mol. Hum. Reprod. 2013, 19, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Guvakova, M.A.; Lee, W.S.Y.; Furstenau, D.K.; Prabakaran, I.; Li, D.C.; Hung, R.; Kushnir, N. The small GTPase Rap1 promotes cell movement rather than stabilizes adhesion in epithelial cells responding to insulin-like growth factor I. Biochem. J. 2014, 463, 257–270. [Google Scholar] [CrossRef]
- Keyes, J.; Ganesan, A.; Molinar-Inglis, O.; Hamidzadeh, A.; Zhang, J.F.; Ling, M.G.; Trejo, J.; Levchenko, A.; Zhang, J. Signaling diversity enabled by Rap1-regulated plasma membrane ERK with distinct temporal dynamics. Elife 2020, 9, e57410. [Google Scholar] [CrossRef]
- Takino, J.I.; Sato, T.; Nagamine, K.; Hori, T. The inhibition of Bax activation-induced apoptosis by RasGRP2 via R-Ras-PI3K-Akt signaling pathway in the endothelial cells. Sci. Rep. 2019, 9, 16717. [Google Scholar] [CrossRef]
- Bromberger, T.; Zhu, L.; Klapproth, S.; Qin, J.; Moser, M. Rap1 and membrane lipids cooperatively recruit talin to trigger integrin activation. J. Cell. Sci. 2019, 132, jcs235531. [Google Scholar] [CrossRef] [PubMed]
- Lilja, J.; Zacharchenko, T.; Georgiadou, M.; Jacquemet, G.; De Franceschi, N.; Peuhu, E.; Hamidi, H.; Pouwels, J.; Martens, V.; Nia, F.H.; et al. SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras. Nat. Cell Biol. 2017, 19, 292–305. [Google Scholar] [CrossRef]
- Lakshmikanthan, S.; Sobczak, M.; Chun, C.; Henschel, A.; Dargatz, J.; Ramchandran, R.; Chrzanowska-Wodnicka, M. Rap1 promotes VEGFR2 activation and angiogenesis by a mechanism involving integrin alphavbeta (3). Blood 2011, 118, 2015–2026. [Google Scholar] [CrossRef]



| Gene Category | Gene Number | Count of PFKM | Functional Description | Reference |
|---|---|---|---|---|
| RPS (Ribosomal protein S2, 3, 5, 6, 7, 8, 10, 11, 12, 14,15,16, 17, 18, 19, 20, 23, 25, 26, 27, 28, 29, 15A, 27A, 4X), RPL (Ribosomal protein L3, 4, 5, 6, 7, 8, 10, 11, 12, 13, 14, 18, 19, 21, 23, 24, 26, 27, 28, 30, 31, 32, 35, 37, 7A, 13A, 18A, 23A, 27A, 35A, P0, P1), LOC102175427 (40S ribosomal protein SA), LOC102186381 (60S ribosomal protein L34), LOC100861018 (60S ribosomal protein L17), FAU(2C ubiquitin like and ribosomal protein S30 fusion), | 61 | 57,458.1 (Total) | Ribosomal genes related to placental development | [29] |
| LOC102177175 (Spleen trypsin inhibitor I), RBP4 (Retinol binding protein), FTH1 (Ferritin heavy chain 1), ACTG1 (Actin gamma 1), SECTM1 (Secreted and transmembrane 1), EEF1A1/G (Eukaryotic translation elongation factor 1 alpha 1/gamma), LOC102178813 (ADP/ATP translocase 3), TMSB4X (Thymosin beta 4 X-linked), SERF2 (Small EDRK-rich factor 2), MYL6 (Myosin light chain 6), LOC102182127 (Clusterin), REXO2 (RNA exonuclease 2), LOC102188434(Cytochrome c oxidase subunit 4 isoform 1), SERPINH1(Serpin family H member 1), PPIB (Peptidylprolyl isomerase B), HSPA8/B1(Heat shock protein family A8/B1) | 18 | 17,987.5 (Total) | Unreported genes in trophoblast transcriptomes | / |
| COL4A1/2, COL18A1 (collagen type IV alpha 1/2 and XVIII alpha 1) | 3 | 708.6/652.8/626.7 | Closely associated with extracellular matrix (ECM) production in placenta | [30,31] |
| CTSB/L (Cathepsin B/L) | 2 | 1750.5/502.5 | Regulated the cell migration, apposition, and remodeled placenta for transport of gases, micronutrients, and macromolecules | [32,33] |
| LOC102184534 (Cystatin-C) | 1 | 1889.1 | Highly expressed in the extravillous trophoblast cells of the basal plate and regulated proteases in placentation. | [32] |
| PAG-8 (Pregnancy-associated glycoprotein-8) | 1 | 1832.6 | Maintenance pregnancy | [34] |
| APOA1 (apolipoprotein A1) | 1 | 1483.6 | Resisted to atherosclerosis and kept cholesterol homeostasis between maternal and fetal | [35] |
| TPT1 (translationally controlled tumor protein 1) | 1 | 1345.3 | Regulated calcium handling transport in trophoblast cells | [36] |
| CALR (Calreticulin) | 1 | 1214.6 | Involved in the invasion of extravillous trophoblasts and syncytialization of villous trophoblasts | [37,38] |
| LOC102188515 (Trophoblast Kunitz domain protein 1-like) | 1 | 1057.3 | Participated in maternal recognition of pregnancy in ruminants | [39,40] |
| LOC102177258 (Placenta-expressed transcript 1 protein) | 1 | 1013.7 | Played a key role in the establishment of a stable trophoblast and endometrial epithelial layers | [41] |
| SOLD1(secreted protein of Ly-6 domain 1) | 1 | 984.15 | Regulated cell invasiveness, placental construction, and development of cotyledonary villi | [42,43,44] |
| RACK1 (receptor for activated C kinase 1) | 1 | 861.1 | Involved in migration and invasion activities in human trophoblast BeWo cell | [45] |
| ANXA2 (annexin A2) | 1 | 802.1 | Impaired decidualization of endometrial stromal cells and promoted embryo implantation and placentation | [46] |
| BSG (Basigin) | 1 | 745.9 | Regulated glycolytic flux to promote cells proliferation in embryo implantation and placental development | [47,48] |
| CLDN4 (Claudin 4) | 1 | 701.5 | Located in multinucleated syncytiotrophoblast layer and participate in modulating trophoblast mobility | [49,50] |
| UBB (Ubiquitin B) | 1 | 608.2 | Involved in the degradation of the extracellular matrix (ECM) and trophoblastic invasion during early pregnancy | [51] |
| P4HB (Prolyl 4-hydroxylase subunit beta) | 1 | 592.2 | Predominantly expressed in human extravillous trophoblasts | [52] |
| CD63 | 1 | 575.1 | Highly expressed in mesenchymal stem cell exosomes in placenta isolated exosomes expressed the exosome markers | [53] |
| LGALS1(Galectin 1) | 1 | 531.7 | Promoted mouse trophoblast stem cell differentiation and potentiates trophoblast fusion in humans | [54,55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, N.; Cheng, W.; Zhou, Y.; Gu, B.; Zhao, Z.; Zhao, Y. Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus). Animals 2021, 11, 2132. https://doi.org/10.3390/ani11072132
Luo N, Cheng W, Zhou Y, Gu B, Zhao Z, Zhao Y. Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus). Animals. 2021; 11(7):2132. https://doi.org/10.3390/ani11072132
Chicago/Turabian StyleLuo, Nanjian, Wenqiang Cheng, Yumei Zhou, Bowen Gu, Zhongquan Zhao, and Yongju Zhao. 2021. "Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus)" Animals 11, no. 7: 2132. https://doi.org/10.3390/ani11072132
APA StyleLuo, N., Cheng, W., Zhou, Y., Gu, B., Zhao, Z., & Zhao, Y. (2021). Screening Candidate Genes Regulating Placental Development from Trophoblast Transcriptome at Early Pregnancy in Dazu Black Goats (Capra hircus). Animals, 11(7), 2132. https://doi.org/10.3390/ani11072132







