Effect of GP19 Peptide Hyperimmune Antiserum on Activated Macrophage during Ehrlichia canis Infection in Canine Macrophage-like Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of GP194-43 Peptide
2.2. Production of Rabbit Antiserum against GP194-43
2.3. Evaluation of Rabbit Antibody against GP194-43 Using ELISA
2.4. Canine Macrophage-Like Cell Lines and Culture Conditions
2.5. E. canis Culture and Stock Conditions
2.6. Infective Inhibition of Ehrlichia canis in DH82 Cell Line
2.6.1. Infective Detection Using Light Microscopy
2.6.2. Infective Detection Using an Immunocytochemistry (ICC) Technique
2.7. Cytokine Gene Expression Profiles
2.8. Data Analysis
3. Results
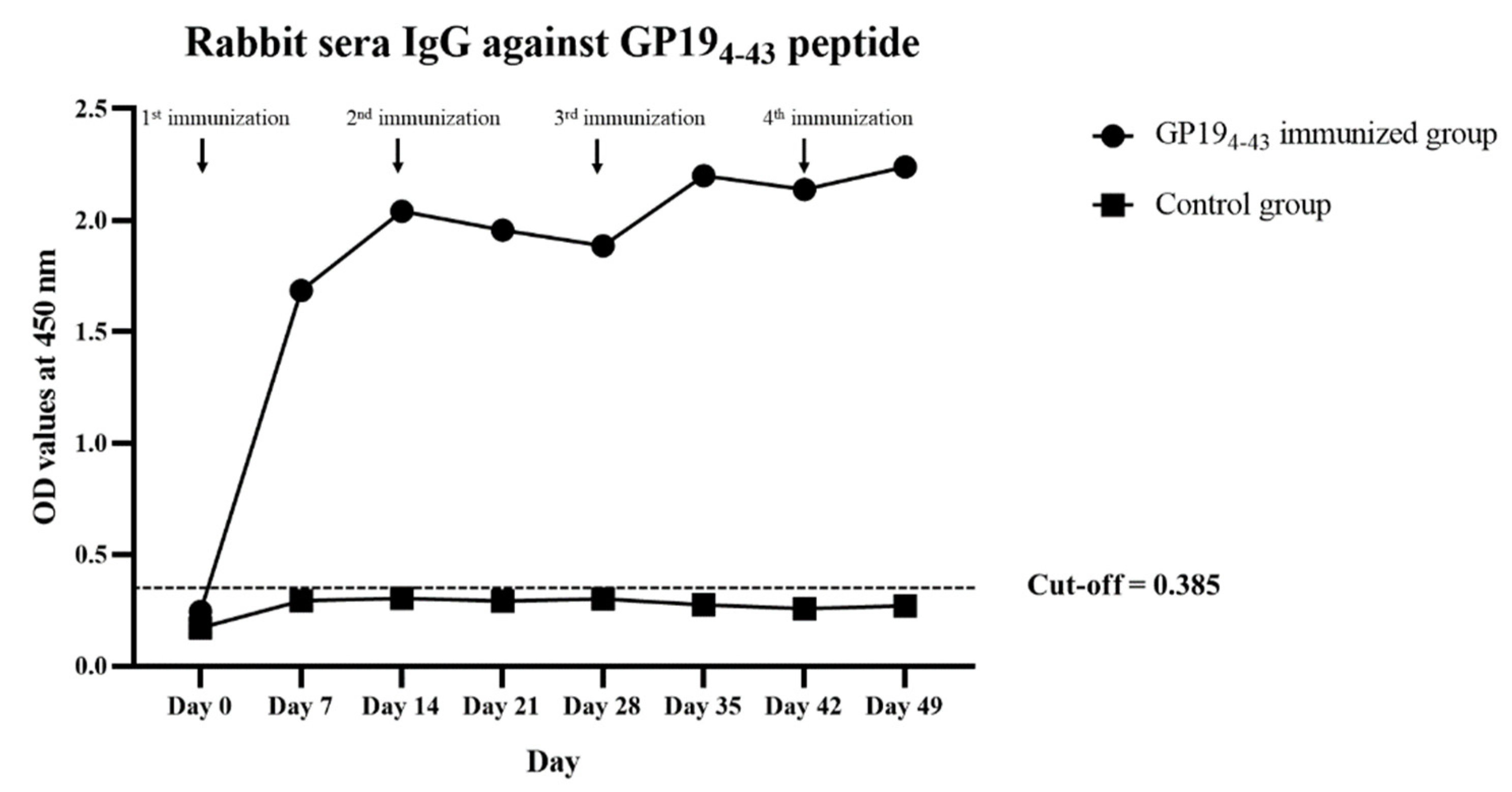
3.1. Rabbit Antibody Response to GP194-43 Peptide
3.2. Infective Inhibition of E. canis to DH82 Cell Line
3.2.1. Positive E. canis Infection Detected by Light Microscopy
3.2.2. E. canis Infective Inhibition Detected by ICC Technique
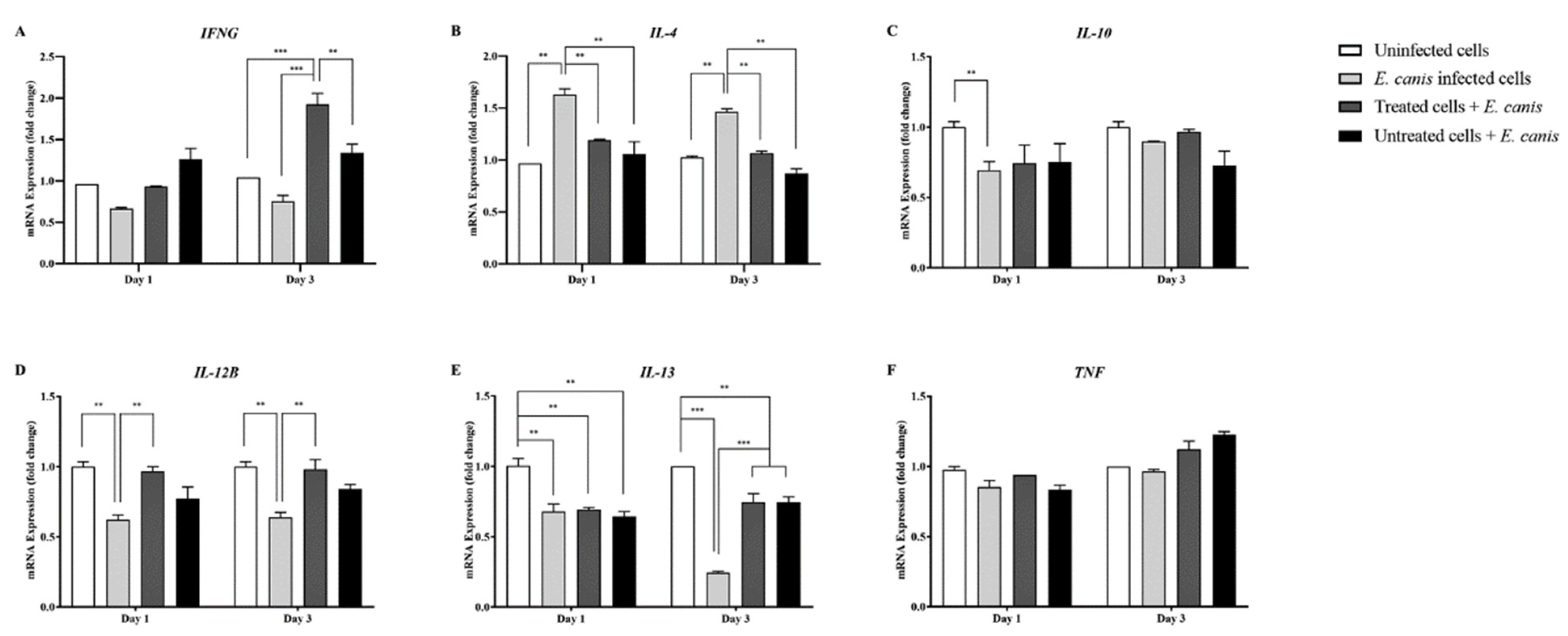
3.3. Relevance of GP194-43 Antiserum on Cytokine Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bremer, W.G.; Schaefer, J.J.; Wagner, E.R.; Ewing, S.A.; Rikihisa, Y.; Needham, G.R.; Jittapalapong, S.; Moore, D.L.; Stich, R.W. Transstadial and intrastadial experimental transmission of Ehrlichia canis by male Rhipicephalus sanguineus. Vet. Parasitol. 2005, 131, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Fourie, J.J.; Stanneck, D.; Luus, H.G.; Beugnet, F.; Wijnveld, M.; Jongejan, F. Transmission of Ehrlichia canis by Rhipicephalus sanguineus ticks feeding on dogs and on artificial membranes. Vet. Parasitol. 2013, 197, 595–603. [Google Scholar] [CrossRef]
- Dumler, J.S.; Barbet, A.F.; Bekker, C.P.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrolho, J.; Simpson, J.; Hawes, P.; Zweygarth, E.; Bell-Sakyi, L. Growth of Ehrlichia canis, the causative agent of canine monocytic ehrlichiosis, in vector and non-vector ixodid tick cell lines. Ticks Tick. Borne. Dis. 2016, 7, 631–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Popov, V.L.; Gao, S.; Walker, D.H.; Yu, X. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 2007, 9, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Harrus, S.; Waner, T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): An overview. Vet. J. 2011, 187, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Waner, T.; Leykin, I.; Shinitsky, M.; Sharabani, E.; Buch, H.; Keysary, A.; Bark, H.; Harrus, S. Detection of platelet-bound antibodies in beagle dogs after artificial infection with Ehrlichia canis. Vet. Immunol. Immunopathol. 2000, 77, 145–150. [Google Scholar] [CrossRef]
- Mylonakis, M.E.; Koutinas, A.F.; Breitschwerdt, E.B.; Hegarty, B.C.; Billinis, C.D.; Leontides, L.S.; Kontos, V.S. Chronic canine ehrlichiosis (Ehrlichia canis): A retrospective study of 19 natural cases. J. Am. Anim. Hosp. Assoc. 2004, 40, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, D.M.; Zhang, X.; Melo, A.L.T.; Pacheco, T.A.; Meneses, A.M.C.; Zanutto, M.S.; Horta, M.C.; Santarém, V.A.; Camargo, L.M.A.; McBride, J.W.; et al. Genetic diversity of Ehrlichia canis in Brazil. Vet. Microbiol. 2013, 164, 315–321. [Google Scholar] [CrossRef]
- Aguirre, E.; Sainz, A.; Dunner, S.; Amusategui, I.; López, L.; Rodríguez-Franco, F.; Luaces, I.; Cortés, O.; Tesouro, M.A. First isolation and molecular characterization of Ehrlichia canis in Spain. Vet. Parasitol. 2004, 125, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Beall, M.J.; Alleman, A.R.; Breitschwerdt, E.B.; Cohn, L.A.; Couto, C.G.; Dryden, M.W.; Guptill, L.C.; Iazbik, C.; Kania, S.A.; Lathan, P.; et al. Seroprevalence of Ehrlichia canis, Ehrlichia chaffeensis and Ehrlichia ewingii in dogs in North America. Parasit. Vectors. 2012, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamani, J.; Lee, C.-C.; Haruna, A.M.; Chung, P.-J.; Weka, P.R.; Chung, Y.-T. First detection and molecular characterization of Ehrlichia canis from dogs in Nigeria. Res. Vet. Sci. 2013, 94, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Nazari, M.; Lim, S.Y.; Watanabe, M.; Sharma, R.S.; Cheng, N.A.; Watanabe, M. Molecular detection of Ehrlichia canis in dogs in Malaysia. PLoS Negl. Trop. Dis. 2013, 7, e1982. [Google Scholar] [CrossRef] [Green Version]
- Perez, M.; Bodor, M.; Zhang, C.; Xiong, Q.; Rikihisa, Y. Human infection with Ehrlichia canis accompanied by clinical signs in Venezuela. Ann. N. Y. Acad. Sci. 2006, 1078, 110–117. [Google Scholar] [CrossRef]
- McBride, J.W.; Walker, D.H. Progress and obstacles in vaccine development for the ehrlichioses. Expert Rev. Vaccines 2010, 9, 1071–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahan, S.; Kelly, P.J.; Mahan, S.M. A preliminary study to evaluate the immune responses induced by immunization of dogs with inactivated Ehrlichia canis organisms. Onderstepoort J. Vet. Res. 2005, 72, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Rudoler, N.; Baneth, G.; Eyal, O.; van Straten, M.; Harrus, S. Evaluation of an attenuated strain of Ehrlichia canis as a vaccine for canine monocytic ehrlichiosis. Vaccine 2012, 31, 226–233. [Google Scholar] [CrossRef]
- Thomas, S.; Thirumalapura, N.R.; Crocquet-Valdes, P.A.; Luxon, B.A.; Walker, D.H. Structure-based vaccines provide protection in a mouse model of ehrlichiosis. PLoS ONE 2011, 6, e27981. [Google Scholar] [CrossRef] [Green Version]
- Dormitzer, P.R.; Ulmer, J.B.; Rappuoli, R. Structure-based antigen design: A strategy for next generation vaccines. Trends Biotechnol. 2008, 26, 659–667. [Google Scholar] [CrossRef]
- Shams, H. Recent developments in veterinary vaccinology. Vet. J. 2005, 170, 289–299. [Google Scholar] [CrossRef]
- Croft, N.P.; Purcell, A.W. Peptidomimetics: Modifying peptides in the pursuit of better vaccines. Expert Rev. Vaccines 2011, 10, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Thirumalapura, N.R.; Crocquet-Valdes, P.A.; Saito, T.B.; Thomas, S.; McBride, J.W.; Walker, D.H. Recombinant Ehrlichia P29 protein induces a protective immune response in a mouse model of ehrlichiosis. Vaccine 2013, 31, 5960–5967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, S. Development of structure-based vaccines for ehrlichiosis. Methods Mol. Biol. 2016, 1403, 519–534. [Google Scholar] [CrossRef]
- Doyle, C.K.; Nethery, K.A.; Popov, V.L.; McBride, J.W. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 2006, 74, 711–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, J.W.; Corstvet, R.E.; Gaunt, S.D.; Boudreaux, C.; Guedry, T.; Walker, D.H. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 2003, 71, 2516–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, J.W.; Doyle, C.K.; Zhang, X.; Cardenas, A.M.; Popov, V.L.; Nethery, K.A.; Woods, M.E. Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 2007, 75, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, Y.-C.; Lee, C.-C.; Tsang, C.-L.; Chung, Y.-T. Detection and characterization of four novel genotypes of Ehrlichia canis from dogs. Vet. Microbiol. 2010, 146, 70–75. [Google Scholar] [CrossRef]
- Nambooppha, B.; Rittipornlertrak, A.; Tattiyapong, M.; Tangtrongsup, S.; Tiwananthagorn, S.; Chung, Y.-T.; Sthitmatee, N. Two different genogroups of Ehrlichia canis from dogs in Thailand using immunodominant protein genes. Infect. Genet. Evol. 2018, 63, 116–125. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, T.; Keysary, A.; Baneth, G.; Miyashiro, S.; Strenger, C.; Waner, T.; McBride, J.W. Genetic and antigenic diversities of major immunoreactive proteins in globally distributed Ehrlichia canis strains. Clin. Vaccine Immunol. 2008, 15, 1080–1088. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, F.; Contioso, V.B.; Stein, V.M.; Carlson, R.; Tipold, A.; Ulrich, R.; Puff, C.; Baumgärtner, W.; Spitzbarth, I. Passage-dependent morphological and phenotypical changes of a canine histiocytic sarcoma cell line (DH82 cells). Vet. Immunol. Immunopathol. 2015, 163, 86–92. [Google Scholar] [CrossRef]
- Keysary, A.; Waner, T.; Strenger, C.; Harrus, S. Cultivation of Ehrlichia canis in a continuous BALB/C mouse macrophage cell culture line. J. Vet. Diagn. Investig. 2001, 13, 521–523. [Google Scholar] [CrossRef] [Green Version]
- Dinarello, C.A. Historical insights into cytokines. Eur. J. Immunol. 2007, 37 (Suppl. S1), S34–S45. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Crowther, J.R. The ELISA Guidebook, 2nd ed.; Humana Press: New York, NY, USA, 2009. [Google Scholar]
- AVMA (American Veterinary Medical Association). AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. Available online: https://www.avma.org/sites/default/files/2020-01/2020_Euthanasia_Final_1-15-20.pdf (accessed on 21 June 2020).
- Teng, C.H.; Palaniappan, R.U.; Chang, Y.F. Cloning and characterization of an Ehrlichia canis gene encoding a protein localized to the morula membrane. Infect. Immun. 2003, 71, 2218–2225. [Google Scholar] [CrossRef] [Green Version]
- Borrathybay, E.; Sawada, T.; Kataoka, Y.; Ohtsu, N.; Takagi, M.; Nakamura, S.; Kawamoto, E. A 39kDa protein mediates adhesion of avian Pasteurella multocida to chicken embryo fibroblast cells. Vet. Microbiol. 2003, 97, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.M.; Gaunt, S.D. Immunocytochemical detection of Ehrlichia platys antigens in canine blood platelets. J. Vet. Diagn. Investig. 1991, 3, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Rizzardi, A.E.; Johnson, A.T.; Vogel, R.I.; Pambuccian, S.E.; Henriksen, J.; Skubitz, A.P.N.; Metzger, G.J.; Schmechel, S.C. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn. Pathol. 2012, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Esteves, I.; Vachiéry, N.; Martinez, D.; Totté, P. Analysis of Ehrlichia ruminantium-specific T1/T2 responses during vaccination with a protective killed vaccine and challenge of goats. Parasite Immunol. 2004, 26, 95–103. [Google Scholar] [CrossRef]
- Cárdenas, A.M.; Doyle, C.K.; Zhang, X.; Nethery, K.; Corstvet, R.E.; Walker, D.H.; McBride, J.W. Enzyme-linked immunosorbent assay with conserved immunoreactive glycoproteins gp36 and gp19 has enhanced sensitivity and provides species-specific immunodiagnosis of Ehrlichia canis infection. Clin. Vac. Immun. 2007, 14, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Chang-Liu, C.M.; Woloschak, G.E. Effect of passage number on cellular response to DNA-damaging agents: Cell survival and gene expression. Cancer Lett. 1997, 113, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Li, J.S.; Yager, E.; Reilly, M.; Freeman, C.; Reddy, G.R.; Reilly, A.A.; Chu, F.K.; Winslow, G.M. Outer membrane protein-specific monoclonal antibodies protect SCID mice from fatal infection by the obligate intracellular bacterial pathogen Ehrlichia chaffeensis. J. Immunol. 2001, 166, 1855–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winslow, G.M.; Yager, E.; Shilo, K.; Volk, E.; Reilly, A.; Chu, F.K. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect. Immun. 2000, 68, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Soong, L.; McBride, J.W.; Valbuena, G.; Olano, J.P.; Feng, H.M.; Walker, D.H. Overproduction of TNF-α by CD8+ Type 1 cells and down-regulation of IFN-γ production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J. Immunol. 2004, 172, 1786–1800. [Google Scholar] [CrossRef] [Green Version]
- Byrom, B.; Obwolo, M.; Barbet, A.F.; Mahan, S.M. A polarized Th1 type immune response to Cowdria Ruminantium infection is detected in immune DBA/2. J. Parasitol. 2000, 86, 983–992. [Google Scholar] [CrossRef]
- Esteves, I.; Bensaid, A.; Martinez, D.; Totté, P. IFN-γ as an indicator of successful immunization of goats vaccinated with a killed Cowdria ruminantium vaccine. Ann. N. Y. Acad. Sci. 2002, 969, 126–130. [Google Scholar] [CrossRef]
- Ismail, N.; Stevenson, H.L.; Walker, D.H. Role of tumor necrosis factor alpha (TNF-α) and interleukin-10 in the pathogenesis of severe murine monocytotropic ehrlichiosis: Increased resistance of TNF receptor p55- and p75-deficient Mice to fatal ehrlichial infection. Infect. Immun. 2006, 74, 1846–1856. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, H.L.; Crossley, E.C.; Thirumalapura, N.; Walker, D.H.; Ismail, N. Regulatory roles of CD1d-restricted NKT cells in the induction of toxic shock-like syndrome in an animal model of fatal ehrlichiosis. Infect. Immun. 2008, 76, 1434–1444. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, H.L.; Estes, M.D.; Thirumalapura, N.R.; Walker, D.H.; Ismail, N. Natural killer cells promote tissue injury and systemic inflammatory responses during fatal Ehrlichia-induced toxic shock-like syndrome. Am. J. Pathol. 2010, 177, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nambooppha, B.; Rittipornlertrak, A.; Muenthaisong, A.; Koonyosying, P.; Tangtrongsup, S.; Tiwananthagorn, S.; Chung, Y.-T.; Sthitmatee, N. Effect of GP19 Peptide Hyperimmune Antiserum on Activated Macrophage during Ehrlichia canis Infection in Canine Macrophage-like Cells. Animals 2021, 11, 2310. https://doi.org/10.3390/ani11082310
Nambooppha B, Rittipornlertrak A, Muenthaisong A, Koonyosying P, Tangtrongsup S, Tiwananthagorn S, Chung Y-T, Sthitmatee N. Effect of GP19 Peptide Hyperimmune Antiserum on Activated Macrophage during Ehrlichia canis Infection in Canine Macrophage-like Cells. Animals. 2021; 11(8):2310. https://doi.org/10.3390/ani11082310
Chicago/Turabian StyleNambooppha, Boondarika, Amarin Rittipornlertrak, Anucha Muenthaisong, Pongpisid Koonyosying, Sahatchai Tangtrongsup, Saruda Tiwananthagorn, Yang-Tsung Chung, and Nattawooti Sthitmatee. 2021. "Effect of GP19 Peptide Hyperimmune Antiserum on Activated Macrophage during Ehrlichia canis Infection in Canine Macrophage-like Cells" Animals 11, no. 8: 2310. https://doi.org/10.3390/ani11082310





