Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Cattle from Central and Southern Chile
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. PCR Screening and STEC Isolation
2.3. Preliminary Virulence Profiling and Molecular Serogrouping
2.4. Biochemical Characterization of STEC Isolates
2.5. Selection of Non-O157 STEC for Full Characterization/WGS
2.6. Saa Gene Typing in STEC Isolated from Cattle
2.7. Antimicrobial Susceptibility
2.8. Whole-Genome Sequencing (WGS)
2.9. Genomic Data Analysis
2.10. Phylogenetic Relatedness among Isolates
2.11. Sequence Accession Numbers
3. Results
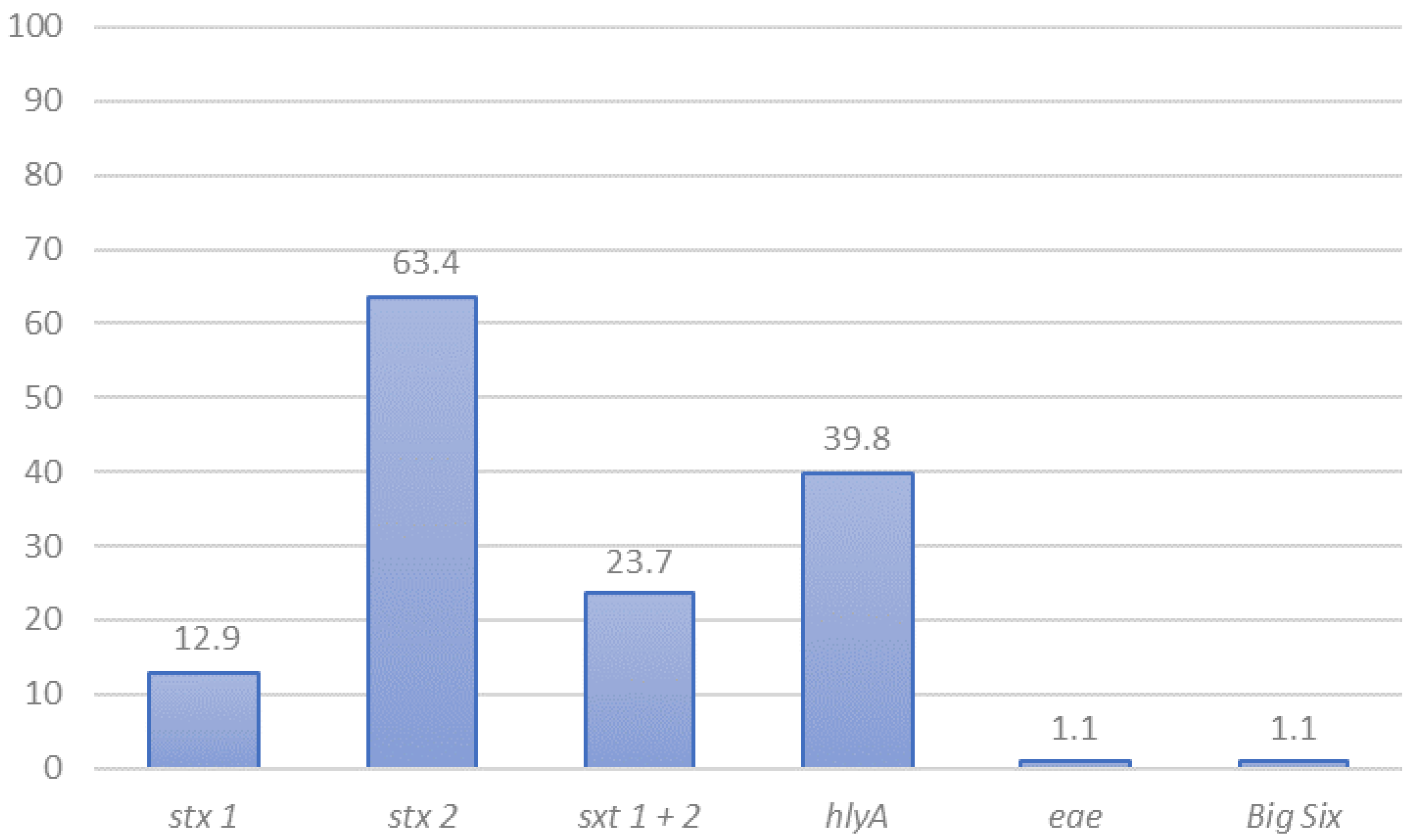
3.1. STEC Detection and Preliminary Characterization by PCR
3.2. Phenotypic Characteristics of STEC Isolated from Cattle
3.3. Antimicrobial Susceptibility
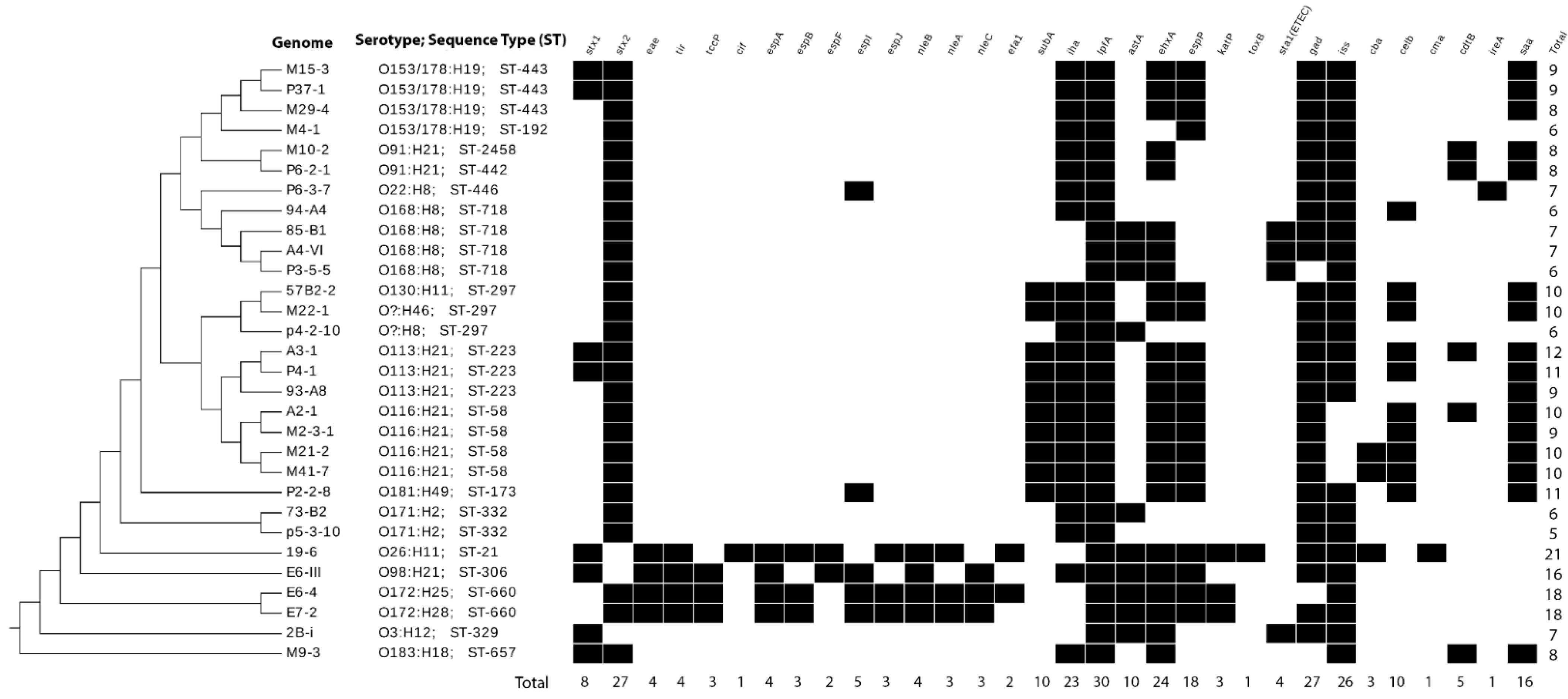
3.4. Sequence Types and Serotypes of Sequenced STEC by Genomic Analyses
3.5. Virulence Genes in STEC Isolated in Chile
3.6. Phylogenetic Relatedness among Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keir, L.S. Shiga toxin associated hemolytic uremic syndrome. Hematol. Oncol. Clin. N. Am. 2015, 29, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Bruyand, M.; Mariani-Kurkdjian, P.; Gouali, M.; de Valk, H.; King, L.A.; Le Hello, S.; Bonacorsi, S.; Loirat, C. Hemolytic uremic syndrome due to Shiga toxin-producing Escherichia coli infection. Med. Mal. Infect. 2018, 48, 167–174. [Google Scholar] [CrossRef]
- FAO/WHO. Hazard identification and characterization: Criteria for categorizing Shiga toxin-producing Escherichia coli on a risk basis. J. Food Prot. 2019, 82, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.; Green, A.; Allen, L.; Ihry, T.; White, P.; Chen, W.S.; Douris, A.; Levine, J. Foodborne outbreaks reported to the U.S. food safety and inspection service, Fiscal Years 2007 through 2012. J. Food Prot. 2016, 79, 442–447. [Google Scholar] [CrossRef]
- BIOHAZ/EFSA. Scientific Opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA J. 2013, 11. [Google Scholar] [CrossRef]
- FAO/WHO. Shiga Toxin-Producing Escherichia coli (STEC) and Food: Attribution, Characterization, and Monitoring: Report; World Health Organization: Rome, Italy, 2018. [Google Scholar]
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Lathrop, S.; Medus, C.; Spina, N.L.; Webb, T.H.; et al. Increased Recognition of Non-O157 Shiga Toxin–Producing Escherichia coli Infections in the United States During 2000–2010: Epidemiologic Features and Comparison with E. coli O157 Infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture (USDA). FSIS. Shiga Toxin-Producing Escherichia coli in Certain Raw Beef Products. Fed. Regist. Available online: https://www.federalregister.gov/documents/2011/11/23/2011-30271/shiga-toxin-producing-escherichia-coli-in-certain-raw-beef-products (accessed on 19 April 2021).
- Intituto de Salud Pública de Chile (ISP). Vigilancia de laboratorio de E. coli productora de toxina Shiga. Chile, 2007–2013. Bol. Inst. Salud Pública Chile 2014, 4, 1–17. [Google Scholar]
- Jay, M.T.; Cooley, M.; Carychao, D.; Wiscomb, G.W.; Sweitzer, R.A.; Crawford-Miksza, L.; Farrar, J.A.; Lau, D.K.; O’Connell, J.; Millington, A.; et al. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, central California coast. Emerg. Infect. Dis. 2007, 13, 1908–1911. [Google Scholar] [CrossRef]
- Söderström, A.; Österberg, P.; Lindqvist, A.; Jönsson, B.; Lindberg, A.; Blide Ulander, S.; Welinder-Olsson, C.; Löfdahl, S.; Kaijser, B.; De Jong, B.; et al. A Large Escherichia coli O157 Outbreak in Sweden Associated with Locally Produced Lettuce. Foodborne Pathog. Dis. 2008, 5, 339–349. [Google Scholar] [CrossRef]
- Wilson, D.; Dolan, G.; Aird, H.; Sorrell, S.; Dallman, T.J.; Jenkins, C.; Robertson, L.; Gorton, R. Farm-to-fork investigation of an outbreak of Shiga toxin-producing Escherichia coli O157. Microb. Genom. 2018. [Google Scholar] [CrossRef]
- Dos Santos, E.C.C.; Castro, V.S.; Cunha-Neto, A.; Dos Santos, L.F.; Vallim, D.C.; Lisbôa, R.D.C.; Carvalho, R.C.T.; Conte, C.A.; Figueiredo, E.E.D.S.; Junior, C.A.C.; et al. Escherichia coli O26 and O113:H21 on carcasses and beef from a slaughterhouse located in Mato Grosso, Brazil. Foodborne Pathog. Dis. 2018, 15, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Heiman, K.E.; Mody, R.K.; Johnson, S.D.; Griffin, P.M.; Gould, L.H. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 2015, 21, 1293–1301. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; van Pelt, W.; van der Voort, M.; Heck, M.; Friesema, I.; Franz, E. Attribution of human infections with Shiga toxin-producing Escherichia coli (STEC) to livestock sources and identification of source-specific risk factors, The Netherlands (2010–2014). Zoonoses Public Health 2018, 65, e8–e22. [Google Scholar] [CrossRef]
- Toro, M.; Rivera, D.; Jiménez, M.F.; Díaz, L.; Navarrete, P.; Reyes-Jara, A. Isolation and characterization of non-O157 Shiga toxin-producing Escherichia coli (STEC) isolated from retail ground beef in Santiago, Chile. Food Microbiol. 2018, 75. [Google Scholar] [CrossRef]
- Li, R.; Tan, X.; Xiao, J.; Wang, H.; Liu, Z.; Zhou, M.; Bi, W.; Miyamoto, T. Molecular screening and characterization of Shiga toxin-producing Escherichia coli in retail foods. Food Control 2016, 60, 180–188. [Google Scholar] [CrossRef]
- Byrne, L.; Vanstone, G.L.; Perry, N.T.; Launders, N.; Adak, G.K.; Godbole, G.; Grant, K.A.; Smith, R.; Jenkins, C. Epidemiology and microbiology of Shiga toxin-producing Escherichia coli other than serogroup O157 in England, 2009-2013. J. Med. Microbiol. 2014, 63, 1181–1188. [Google Scholar] [CrossRef]
- McWilliams, B.D.; Torres, A.G. Enterohemorrhagic Escherichia coli Adhesins. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Franz, E.; Delaquis, P.; Morabito, S.; Beutin, L.; Gobius, K.; Rasko, D.A.; Bono, J.; French, N.; Osek, J.; Lindstedt, B.A.; et al. Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. Int. J. Food Microbiol. 2014, 187, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Stromberg, Z.R.; Lewis, G.L.; Marx, D.B.; Moxley, R.A. Comparison of enrichment broths for supporting growth of Shiga Toxin-Producing Escherichia coli. Curr. Microbiol. 2015, 71, 214–219. [Google Scholar] [CrossRef]
- Toro, M.; Najjar, M.B.; Ju, W.; Brown, E.; Zhao, S.; Meng, J. Molecular serogrouping of Shiga toxin-producing Escherichia coli using suspension array. Foodborne Pathog. Dis. 2013, 10. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Woodrow, M.C.; Doyle, R.M.; Lanser, J.A.; Paton, J.C. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 1999, 37, 3357–3361. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Griffiths, M.W. PCR differentiation of Escherichia coli from other Gram-negative bacteria using primers derived from the nucleotide sequences flanking the gene encoding the universal stress protein. Lett. Appl. Microbiol. 1998, 27, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Fratamico, P.M.; Strobaugh, T.P. Evaluation of an enzyme-linked immunosorbent assay, direct immunofluorescent filter technique, and multiplex polymerase chain reaction for detection of Escherichia coli O157:H7 seeded in beef carcass wash water. J. Food Prot. 1998, 61, 934–938. [Google Scholar] [CrossRef]
- Xia, X.; Meng, J.; McDermott, P.F.; Ayers, S.; Blickenstaff, K.; Tran, T.T.; Abbott, J.; Zheng, J.; Zhao, S. Presence and characterization of Shiga toxin-producing Escherichia coli and other potentially diarrheagenic E. coli strains in retail meats. Appl. Environ. Microbiol. 2010, 76, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Miko, A.; Rivas, M.; Bentancor, A.; Delannoy, S.; Fach, P.; Beutin, L. Emerging types of Shiga toxin-producing E. coli (STEC) O178 present in cattle, deer, and humans from Argentina and Germany. Front. Cell. Infect. Microbiol. 2014, 4, 78. [Google Scholar] [CrossRef]
- Rivas, M.; Miliwebsky, E.; Deza, N. Manual de Procedimientos Diagnóstico y Caracterización de Escherichia coli O157 Productor de Toxina Shiga a Partir de Especimenes Clínicos; Instituto Nacional de Enfermedades Infecciosas: Buenos Aires, Argentina, 2007; pp. 1–52. [Google Scholar]
- Verhaegen, B.; De Reu, K.; Heyndrickx, M.; De Zutter, L. Comparison of six chromogenic agar media for the isolation of a broad variety of non-o157 Shigatoxin-producing Escherichia coli (STEC) Serogroups. Int. J. Environ. Res. Public Health 2015, 12, 6965–6978. [Google Scholar] [CrossRef]
- Beutin, L.; Montenegro, M.A.; Orskov, I.; Orskov, F.; Prada, J.; Zimmermann, S.; Stephan, R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 1989, 27, 2559–2564. [Google Scholar] [CrossRef]
- Lucchesi, P.M.A.; Krüger, A.; Parma, A.E. Distribution of saa gene variants in verocytotoxigenic Escherichia coli isolated from cattle and food. Res. Microbiol. 2006, 157, 263–266. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. 2015. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 19 April 2021).
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2015. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Escalona, N.; Toro, M.; Rump, L.V.; Cao, G.; Nagaraja, T.G.; Meng, J. Virulence gene profiles and clonal relationships of Escherichia coli O26:H11 isolates from feedlot cattle as determined by whole-genome sequencing. Appl. Environ. Microbiol. 2016, 82, 3900–3912. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Ylinen, E.; Salmenlinna, S.; Halkilahti, J.; Jahnukainen, T.; Korhonen, L.; Virkkala, T.; Rimhanen-Finne, R.; Nuutinen, M.; Kataja, J.; Arikoski, P.; et al. Hemolytic uremic syndrome caused by Shiga toxin-producing Escherichia coli in children: Incidence, risk factors, and clinical outcome. Pediatr. Nephrol. 2020. [Google Scholar] [CrossRef]
- Petro, C.D.; Trojnar, E.; Sinclair, J.; Liu, Z.-M.; Smith, M.; O’Brien, A.D.; Melton-Celsa, A. Shiga toxin type 1a (Stx1a) reduces the toxicity of the more potent Stx2a in vivo and in vitro. Infect. Immun. 2019, 87, e00787-18. [Google Scholar] [CrossRef]
- Borie, C.F.; Monreal, Z.; Martinez, J.; Arellano, C.; Prado, V. Detection and characterization of enterohaemorrhagic Escherichia coli in slaughtered cattle. J. Vet. Med. Ser. A Physiol. Pathol. Clin. Med. 1997, 44, 273–279. [Google Scholar] [CrossRef]
- Fernández, D.; Rodríguez, E.M.; Arroyo, G.H.; Padola, N.L.; Parma, A.E. Seasonal variation of Shiga toxin-encoding genes (stx) and detection of E. coli O157 in dairy cattle from Argentina. J. Appl. Microbiol. 2009, 106, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Rhades, L.C.; Larzábal, M.; Bentancor, A.; García, J.S.Y.; Babinec, F.J.; Cataldi, A.; Amigo, N.; Baldone, V.N.; Urquiza, L.; Delicia, P.J.; et al. A one-year longitudinal study of enterohemorrhagic Escherichia coli O157 fecal shedding in a beef cattle herd. Res. Vet. Sci. 2019, 127, 27–32. [Google Scholar] [CrossRef]
- Fernández, D.; Irino, K.; Sanz, M.E.; Padola, N.L.; Parma, A.E. Characterization of Shiga toxin-producing Escherichia coli isolated from dairy cows in Argentina. Lett. Appl. Microbiol. 2010, 51, 377–382. [Google Scholar] [CrossRef]
- Blanco, M.; Blanco, J.E.; Mora, A.; Rey, J.; Alonso, J.M.; Hermoso, M.; Hermoso, J.; Alonso, M.P.; Dahbi, G.; González, E.A.; et al. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 2003, 41, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Hornitzky, M.A.; Vanselow, B.A.; Walker, K.; Bettelheim, K.A.; Corney, B.; Gill, P.; Bailey, G.; Djordjevic, S.P. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian cattle. Appl. Environ. Microbiol. 2002, 68, 6439. [Google Scholar] [CrossRef]
- Bugarel, M.; Beutin, L.; Scheutz, F.; Loukiadis, E.; Fach, P. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl. Environ. Microbiol. 2011, 77, 2275–2281. [Google Scholar] [CrossRef]
- Beutin, L.; Krause, G.; Zimmermann, S.; Kaulfuss, S.; Gleier, K. Characterization of Shiga Toxin-Producing Escherichia coli Strains Isolated from Human Patients in Germany over a 3-Year Period. J. Clin. Microbiol. 2004, 42, 1099–1108. [Google Scholar] [CrossRef]
- Sanso, A.M.; Bustamante, A.V.; Krüger, A.; Cadona, J.S.; Alfaro, R.; Cáceres, M.E.; Fernández, D.; Lucchesi, P.M.A.; Padola, N.L. Molecular epidemiology of Shiga toxin-producing O113:H21 isolates from cattle and meat. Zoonoses Public Health 2018, 65, 569–577. [Google Scholar] [CrossRef]
- Cadona, J.S.; Bustamante, A.V.; González, J.; Sanso, A.M. Genetic relatedness and novel sequence types of non-O157 Shiga toxin-producing Escherichia coli Strains Isolated in Argentina. Front. Cell. Infect. Microbiol. 2016, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.; Padola, N.L.; Krüger, A.; Sanz, M.E.; Blanco, J.E.; González, E.A.; Dahbi, G.; Mora, A.; Bernárdez, M.I.; Etcheverría, A.I.; et al. Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2004, 7, 269–276. [Google Scholar]
- Rivelli Zea, S.M.; Padola, N.L.; Etcheverría, A.I.; Florentín, M.; Acuña, P.; Rodríguez, F.; Colello, R.; Guillén Fretes, R.M. Caracterización molecular de aislamientos de Escherichia coli productores de toxina Shiga obtenidos en 2 establecimientos ganaderos del Paraguay. Rev. Argent. Microbiol. 2020, 52, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, B.; Yambao, J.C.; Lee, B.G. Draft Genome Sequences of Escherichia coli O113:H21 Strains Recovered from a Major Produce Production Region in California. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Mellmann, A.; Bielaszewska, M.; Köck, R.; Friedrich, A.W.; Fruth, A.; Middendorf, B.; Harmsen, D.; Schmidt, M.A.; Karch, H. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 2008, 14, 1287–1290. [Google Scholar] [CrossRef]
- Käppeli, U.; Hächler, H.; Giezendanner, N.; Beutin, L.; Stephan, R. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerg. Infect. Dis. 2011, 17, 180–185. [Google Scholar] [CrossRef]
- Hines, J.Z.; Bancroft, J.; Powell, M.; Hedberg, K. Case Finding Using Syndromic Surveillance Data During an Outbreak of Shiga Toxin-Producing Escherichia coli O26 Infections, Oregon, 2015. Public Health Rep. 2017, 132, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Scavia, G.; Gianviti, A.; Labriola, V.; Chiani, P.; Maugliani, A.; Michelacci, V.; Minelli, F.; Tozzoli, R.; Caprioli, A.; Morabito, S. A case of haemolytic uraemic syndrome (HUS) revealed an outbreak of Shiga toxin-2-producing Escherichia coli O26:H11 infection in a nursery, with long-lasting shedders and person-to-person transmission, Italy 2015. J. Med. Microbiol. 2018, 67, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086. [Google Scholar] [CrossRef]
- Zhou, M.; Ding, X.; Ma, F.; Xu, Y.; Zhang, J.; Zhu, G.; Lu, Y. Long polar fimbriae contribute to pathogenic Escherichia coli infection to host cells. Appl. Microbiol. Biotechnol. 2019, 103, 7317–7324. [Google Scholar] [CrossRef]
- Vergara, A.F.; Vidal, R.M.; Torres, A.G.; Farfan, M.J. Long polar fimbriae participates in the induction of neutrophils transepithelial migration across intestinal cells infected with enterohemorrhagic E. coli O157:H7. Front. Cell. Infect. Microbiol. 2015, 4, 185. [Google Scholar] [CrossRef]
- Bosilevac, J.M.; Koohmaraie, M. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 2011, 77, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Srimanote, P.; Woodrow, M.C.; Paton, J.C. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 2001, 69, 6999–7009. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.; Chinen, I.; Guth, B.E.C. Enterohemorrhagic (Shiga toxin- producing) Escherichia coli. In Escherichia coli in the Americas; Torres, A.G., Ed.; Springer: Cham, Switzerland, 2016; pp. 97–123. ISBN 978-3-319-45092-6. [Google Scholar]
- Freedman, S.B.; Xie, J.; Neufeld, M.S.; Hamilton, W.L.; Hartling, L.; Tarr, P.I. Shiga toxin-producing Escherichia coli infection, antibiotics, and risk of developing hemolytic uremic syndrome: A meta-analysis. Clin. Infect. Dis. 2016. [Google Scholar] [CrossRef]
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Begum, J.; Mir, N.A.; Dev, K.; Khan, I.A. Dynamics of antibiotic resistance with special reference to Shiga toxin-producing Escherichia coli infections. J. Appl. Microbiol. 2018, 125, 1228–1237. [Google Scholar] [CrossRef]
- Cergole-Novella, M.C.; Campos Pignatari, A.C.; Castanheira, M.; Cabilio Guth, B.E. Molecular typing of antimicrobial-resistant Shiga-toxin-producing Escherichia coli strains (STEC) in Brazil. Res. Microbiol. 2011, 162, 117–123. [Google Scholar] [CrossRef]
- Sow, A.G.; Wane, A.A.; Diallo, M.H.; Boye, C.S.-B.; Aïdara-Kane, A. Genotypic characterization of antibiotic-resistant Salmonella Enteritidis isolates in Dakar, Senegal. J. Infect. Dev. Ctries. 2007, 1, 284–288. [Google Scholar]
- Jenkins, C.; Perry, N.T.; Cheasty, T.; Shaw, D.J.; Frankel, G.; Dougan, G.; Gunn, G.J.; Smith, H.R.; Paton, A.W.; Paton, J.C. Distribution of the saa gene in strains of Shiga toxin-producing Escherichia coli of human and bovine origins. J. Clin. Microbiol. 2003, 41, 1775–1778. [Google Scholar] [CrossRef][Green Version]
- Colello, R.; Krüger, A.; Velez, M.V.; Del Canto, F.; Etcheverría, A.I.; Vidal, R.; Padola, N.L. Identification and detection of iha subtypes in LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from humans, cattle and food. Heliyon 2019, 5, e03015. [Google Scholar] [CrossRef]
- Ekong, P.S.; Sanderson, M.W.; Bello, N.M.; Noll, L.W.; Cernicchiaro, N.; Renter, D.G.; Bai, J.; Nagaraja, T.G. Bayesian estimation of true prevalence, sensitivity and specificity of three diagnostic tests for detection of Escherichia coli O157 in cattle feces. Prev. Vet. Med. 2017, 148, 21–27. [Google Scholar] [CrossRef]
- Arancia, S.; Iurescia, M.; Lorenzetti, S.; Stravino, F.; Buccella, C.; Caprioli, A.; Franco, A.; Battisti, A.; Morabito, S.; Tozzoli, R. Detection and isolation of Shiga Toxin-producing Escherichia coli (STEC) strains in caecal samples from pigs at slaughter in Italy. Vet. Med. Sci. 2019, 5, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Parsons, B.D.; Zelyas, N.; Berenger, B.M.; Chui, L. Detection, characterization, and typing of Shiga toxin-producing Escherichia coli. Front. Microbiol. 2016, 7, 478. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tian, S.; Thaker, H.; Dong, M. Shiga Toxins: An Update on Host Factors and Biomedical Applications. Toxins 2021, 13, 222. [Google Scholar] [CrossRef] [PubMed]
- Tajeddin, E.; Ganji, L.; Hasani, Z.; Ghoalm Mostafaei, F.S.; Azimirad, M.; Torabi, P.; Mohebbi, S.R.; Aghili, N.; Gouya, M.M.; Eshrati, B.; et al. Shiga toxin-producing bacteria as emerging enteric pathogens associated with outbreaks of foodborne illness in the Islamic Republic of Iran. East Mediterr. Health J. 2020, 26, 976–981. [Google Scholar] [CrossRef]


| Type of PCR | Target | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Annealing Temp (°C) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|---|
| Multiplex or Singleplex | stx1 | CAGTTAATGTGGTGGCGAAGG | CACCAGACAATGTAACCGCTG | 56 | 348 | [22,23] |
| stx2 | ATCCTATTCCCGGGAGTTTACG | GCGTCATCGTATACACAGGAGC | 584 | |||
| Singleplex | E. coli uspA | CCGATACGCTGCCAATCAGT | ACGCAGACCGTAGGCCAGAT | 58 | 884 | [24] |
| Singleplex | eae | ATTACCATCCACACAGACGGT | ACAGCGTGGTTGGATCAACCT | 63 | 397 | [25] |
| Singleplex | hlyA | AGCCGGAACAGTTCTCTCAG | CCAGCATAACAGCCGATGT | 60 | 526 | [26] |
| Multiplex or Singleplex | O26 wzx | GTGTGTCTGGTTCGTATTTTTTATCTG | CCTTATATCCCAATATAGTACCCACCC | 56 | 438 | [22] |
| O45 wzx | GGTCGATAACTGGTATGCAATATG | CTAGGCAGAAAGCTATCAACCAC | 341 | |||
| O103 wzy | TTATACAAATGGCGTGGATTGGAG | TGCAGACACATGAAAAGTTGATGC | 385 | |||
| O111 wzx | TTCGATGTTGCGAGGAATAATTC | GTGAGAGCCCACCAGTTAATTGAAG | 362 | |||
| O121 wzy | AGTGGGGAAGGGCGTTACTTATC | CAATGAGTGCAGGCAAAATGGAG | 366 | |||
| O145 wzy | CCTGTCTGTTGCTTCAGCCCTTT | CTGTGCGCGAACCACTGCTAAT | 392 |
| Isolate Name | CFSAN Number | Accession Number (SRA) | Location * | Sequence Type | Serotype | stx Gene Subtype | eae Gene Subtype |
|---|---|---|---|---|---|---|---|
| M22-1 | CFSAN066373 | SRX3735307 | Southern | 297 | O?:H46 | 2 | - |
| p4-2-10 | CFSAN066356 | SRX3735281 | Central | 297 | O?:H8 | 2 | - |
| A3-1 | CFSAN066341 | SRX3735341 | Central | 223 | O113:H21 | 1, 2 | - |
| 93-A8 | CFSAN066380 | SRX3735290 | Southern | 223 | O113:H21 | 2a | - |
| P4-1. | CFSAN066382 | SRX3735276 | Central | 223 | O113:H21 | 1a, 2a | - |
| A2-1 | CFSAN066340 | SRX3735340 | Central | 58 | O116:H21 | 2a | - |
| M21-2 | CFSAN066372 | SRX3735329 | Southern | 58 | O116:H21 | 2 | - |
| M2-3-1 | CFSAN066391 | SRX3735309 | Southern | 58 | O116:H21 | 2a | - |
| M41-7 | CFSAN066376 | SRX3735286 | Southern | 58 | O116:H21 | 2 | - |
| 57-B2-2 | CFSAN066390 | SRX3735310 | Southern | 297 | O130:H11 | 2 | - |
| M4-1 | CFSAN066365 | SRX3735322 | Southern | 192 | O153/178:H19 | 2 | - |
| M15-3 | CFSAN066370 | SRX3735321 | Southern | 443 | O153/178:H19 | 1, 2 | - |
| M29-4 | CFSAN066375 | SRX3735287 | Southern | 443 | O153/178:H19 | 2c | - |
| P37-1 | CFSAN066386 | SRX3735308 | Central | 443 | O153/178:H19 | 1a, 2a | - |
| A4-VI | CFSAN066342 | SRX3735338 | Central | 718 | O168:H8 | 2g | - |
| P3-5-5 | CFSAN066355 | SRX3735282 | Central | 718 | O168:H8 | 2g | - |
| 85-B1 | CFSAN066379 | SRX3735291 | Southern | 718 | O168:H8 | 2g | - |
| 94-A4 | CFSAN066381 | SRX3735277 | Southern | 718 | O168:H8 | 2 | - |
| p5-3-10 | CFSAN066357 | SRX3735280 | Central | 332 | O171:H2 | 2a | - |
| 73-B2 | CFSAN066378 | SRX3735288 | Southern | 332 | O171:H2 | 2c | - |
| E6-4 | CFSAN066346 | SRX3735347 | Central | 660 | O172:H25 | 2a | Epsilon-3 |
| E7-2 | CFSAN066349 | SRX3735285 | Central | 660 | O172:H28 | 2a | Epsilon-3 |
| P2-2-8 | CFSAN066354 | SRX3735283 | Central | 173 | O181:H49 | 2c | - |
| M9-3 | CFSAN066366 | SRX3735323 | Southern | 657 | O183:H18 | 1, 2 | - |
| P6-3-7 | CFSAN066360 | SRX3735293 | Central | 446 | O22:H8 | 2c | - |
| 19-6 | CFSAN066388 | SRX3735346 | Southern | 21 | O26:H11 | 1a | Beta-1 |
| 2B-i | CFSAN066353 | SRX3735284 | Southern | 329 | O3:H12 | 1a | - |
| M10-2 | CFSAN066367 | SRX3735324 | Southern | 2458 | O91:H21 | 2a | - |
| P6-2-1 | CFSAN066358 | SRX3735279 | Central | 442 | O91:H21 | 2a | - |
| E6-III | CFSAN066345 | SRX3735339 | Central | 306 | O98:H21 | 1a | Alpha-6 |
| Location | Number of Farms | Isolation (%) |
|---|---|---|
| Central | 14 | 39/155 (25.2) |
| Southern | 5 + 1 slaughterhouse | 17/291 (5.8) |
| Total | 20 | 56/446 (12.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz, L.; Gutierrez, S.; Moreno-Switt, A.I.; Hervé, L.P.; Hamilton-West, C.; Padola, N.L.; Navarrete, P.; Reyes-Jara, A.; Meng, J.; González-Escalona, N.; et al. Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Cattle from Central and Southern Chile. Animals 2021, 11, 2388. https://doi.org/10.3390/ani11082388
Díaz L, Gutierrez S, Moreno-Switt AI, Hervé LP, Hamilton-West C, Padola NL, Navarrete P, Reyes-Jara A, Meng J, González-Escalona N, et al. Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Cattle from Central and Southern Chile. Animals. 2021; 11(8):2388. https://doi.org/10.3390/ani11082388
Chicago/Turabian StyleDíaz, Leonela, Sebastian Gutierrez, Andrea I Moreno-Switt, Luis Pablo Hervé, Christopher Hamilton-West, Nora Lía Padola, Paola Navarrete, Angélica Reyes-Jara, Jianghong Meng, Narjol González-Escalona, and et al. 2021. "Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Cattle from Central and Southern Chile" Animals 11, no. 8: 2388. https://doi.org/10.3390/ani11082388
APA StyleDíaz, L., Gutierrez, S., Moreno-Switt, A. I., Hervé, L. P., Hamilton-West, C., Padola, N. L., Navarrete, P., Reyes-Jara, A., Meng, J., González-Escalona, N., & Toro, M. (2021). Diversity of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Cattle from Central and Southern Chile. Animals, 11(8), 2388. https://doi.org/10.3390/ani11082388










