Resilience of Faecal Microbiota in Stabled Thoroughbred Horses Following Abrupt Dietary Transition between Freshly Cut Pasture and Three Forage-Based Diets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design and Trial Management
Data Recording and Sample Collection
2.3. DNA Extraction, Amplicon Library Construction and Sequencing
2.4. Bioinformatics and Statistical Analyses
3. Results
3.1. Animal Health Monitoring
3.2. Population Dynamics of the Faecal Bacterial Community
Metrics of Sequencing Data and Rarefaction Analysis
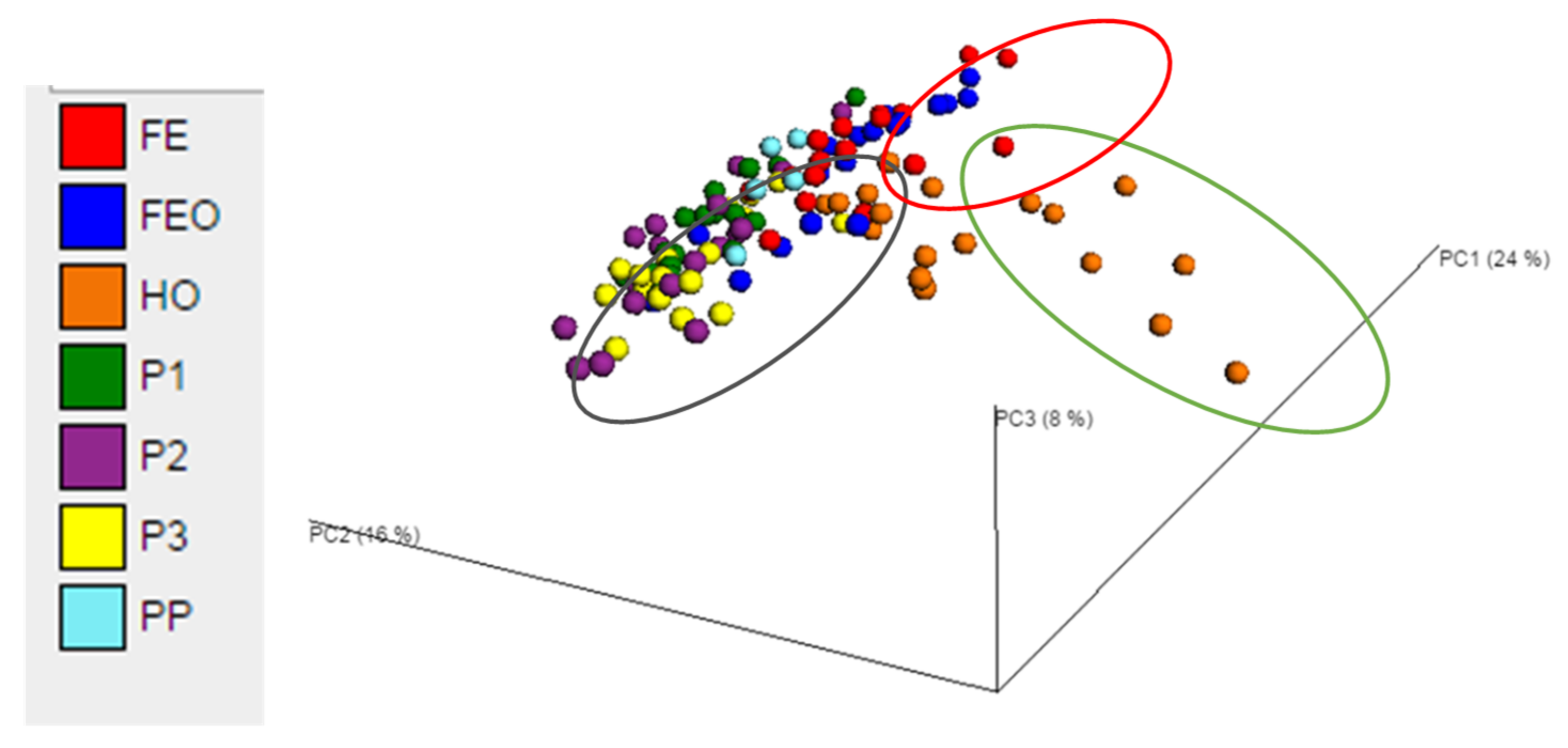
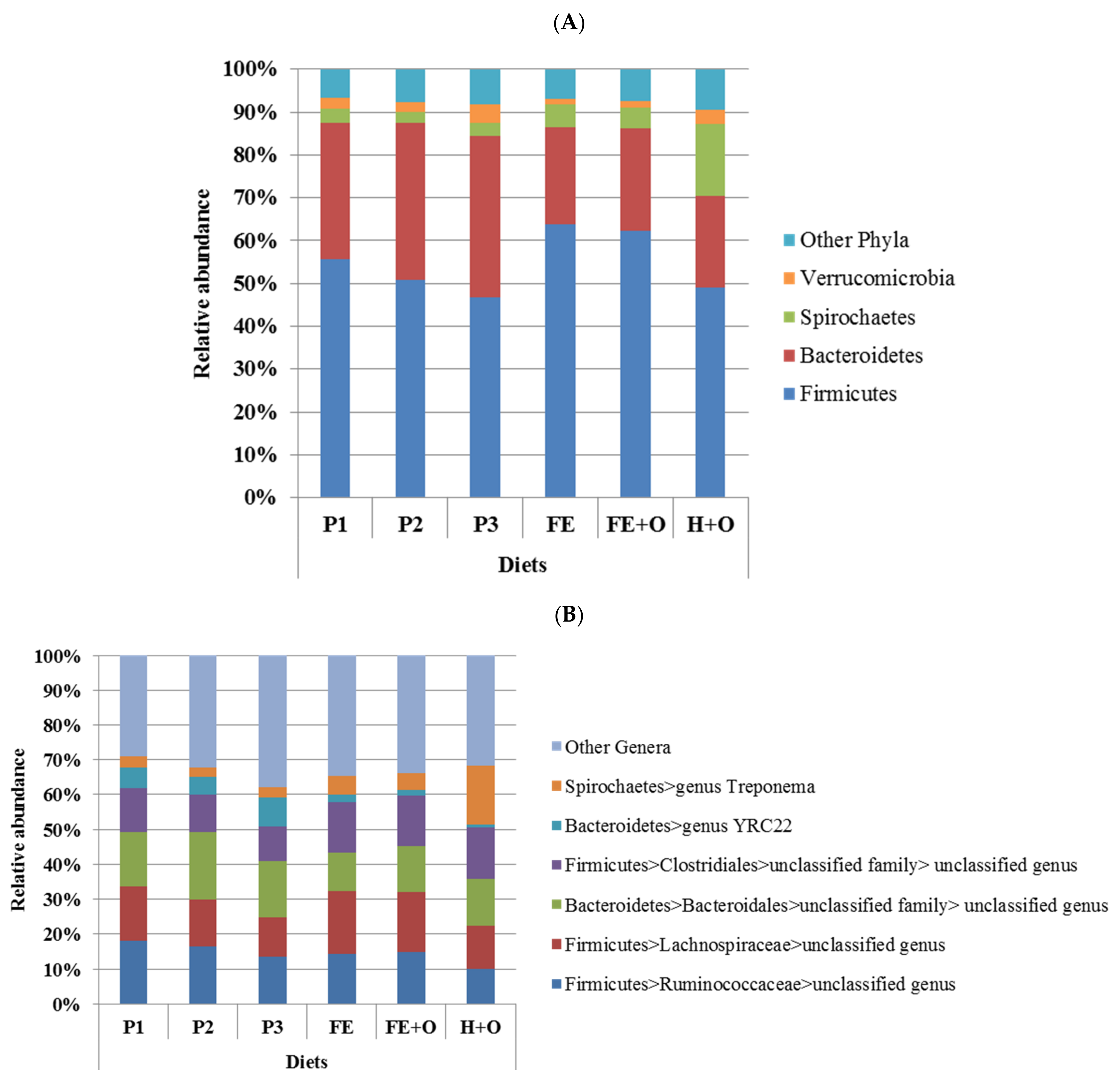
3.3. Effects of Diet on the Faecal Microbiota
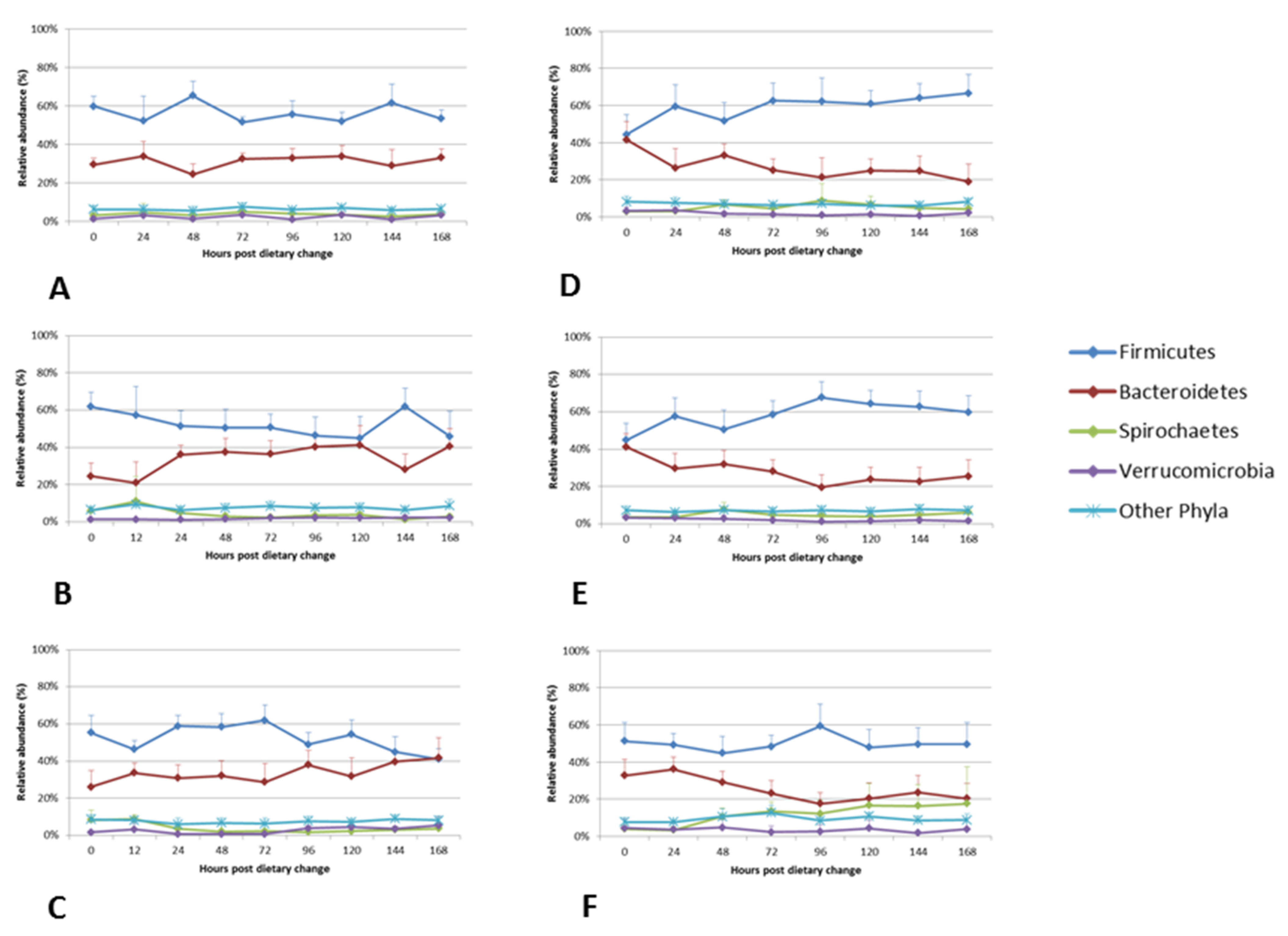
3.4. Dynamics and Stability of the Faecal Bacterial Community Following Dietary Change
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janis, C. The evolutionary strategy of the Equidae and the origins of rumen and cecal digestion. Evolution 1976, 30, 757–774. [Google Scholar] [CrossRef]
- Glinsky, M.; Smith, R.; Spires, H.; Davis, C. Measurement of volatile fatty acid production rates in the cecum of the pony. J. Anim. Sci. 1976, 42, 1465–1470. [Google Scholar] [CrossRef] [Green Version]
- Lewis, L.D.; Knight, A.; Lewis, B.; Lewis, C. Equine Clinical Nutrition: Feeding and Care; Wiley-Blackwell: Baltimore, MD, USA, 1995; 587p. [Google Scholar]
- National Research Council. Nutrient Requirements of Horses, 6th ed.; National Academies Press: Washington, DC, USA, 2007; 357p. [Google Scholar]
- Fernandes, K.A.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Thomas, D.G. A cross-sectional survey of rider and horse demographics, and the feeding, health and management of Pony Club horses in New Zealand. Proc. N. Z. Soc. Anim. Prod. 2014, 74, 11–16. [Google Scholar]
- Verhaar, N.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Rosanowski, S.M. The feeding practices and estimated workload in a cohort of New Zealand competition horses. J. Equine Vet. Sci. 2014, 34, 1257–1262. [Google Scholar] [CrossRef]
- Fernandes, K.A.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Thomas, D.G. Body condition and morphometric measures of adiposity in a cohort of Pony Club horses and ponies in New Zealand. Proc. N. Z. Soc. Anim. Prod. 2015, 75, 195–199. [Google Scholar]
- Rogers, C.W.; Gee, E.K.; Firth, E.C. A cross-sectional survey of Thoroughbred stud farm management in the North Island of New Zealand. N. Z. Vet. J. 2007, 55, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Williamson, A.; Rogers, C.W.; Firth, E.C. A survey of feeding, management and faecal pH of Thoroughbred racehorses in the North Island of New Zealand. N. Z. Vet. J. 2007, 55, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, M.; Hoskin, S.O.; Rogers, C.W.; Grinberg, A. Fecal pH and microbial populations in Thoroughbred horses during transition from pasture to concentrate feeding. J. Equine Vet. Sci. 2013, 33, 215–222. [Google Scholar] [CrossRef]
- Cohen, N.; Gibbs, P.; Woods, A. Dietary and other management factors associated with colic in horses. J. Am. Vet. Med. Assoc. 1999, 215, 53–60. [Google Scholar] [PubMed]
- Garber, A.; Hastie, P.; McGuinness, D.; Malarange, P.; Murray, J.A. Abrupt dietary changes between grass and hay alter faecal microbiota of ponies. PLoS ONE 2020, 15, e0237869. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.C.; Weese, J.S. The equine intestinal microbiome. Anim. Health Res. Rev. 2012, 13, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, K.A.; Kittelmann, S.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Bermingham, E.N.; Thomas, D.G. Faecal microbiota of forage-fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS ONE 2014, 9, e112846. [Google Scholar] [CrossRef] [Green Version]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Newbold, C.J. Identification of a core bacterial community within the large intestine of the horse. PLoS ONE 2013, 8, e77660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, B.E.; Dehority, B.A. Effects of diet and hindgut defaunation on diet digestibility and microbial concentrations in the cecum and colon of the horse. J. Anim. Sci. 1993, 71, 3350–3358. [Google Scholar] [CrossRef] [Green Version]
- Harlow, B.E.; Lawrence, L.M.; Hayes, S.H.; Crum, A.; Flythe, M.D. Effect of dietary starch source and concentration on equine fecal microbiota. PLoS ONE 2016, 11, e0154037. [Google Scholar] [CrossRef] [Green Version]
- Destrez, A.; Grimm, P.; Cézilly, F.; Julliand, V. Changes of the hindgut microbiota due to high-starch diet can be associated with behavioral stress response in horses. Physiol. Behav. 2015, 149, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.C.; Avershina, E.; Mydland, L.T.; Næsset, J.A.; Austbø, D.; Moen, B.; Måge, I.; Rudi, K. High nutrient availability reduces the diversity and stability of the equine caecal microbiota. Microb. Ecol. Health Dis. 2015, 26, 27216. [Google Scholar] [CrossRef]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Brit. J. Nutr. 2012, 107, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Kristoffersen, C.T. Diet Effects on the Short-Term Temporal Dynamics of the Equine Hindgut Microbiota. Master’s Thesis, Norwegian University of Life Sciences, As, Norway, 2014. [Google Scholar]
- Warzecha, C.M.; Coverdale, J.A.; Janecka, J.E.; Leatherwood, J.L.; Pinchak, W.E.; Wickersham, T.A.; McCann, J.C. Influence of short-term dietary starch inclusion on the equine cecal microbiome. J. Anim. Sci. 2017, 95, 5077–5090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Jassim, R.A.M.; Andrews, F.M. The bacterial community of the horse gastrointestinal tract and its relation to fermentative acidosis, laminitis, colic, and stomach ulcers. Vet. Clin. N. Am. Equine 2009, 25, 199–215. [Google Scholar] [CrossRef]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stämpfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef] [Green Version]
- Milinovich, G.J.; Trott, D.J.; Burrell, P.C.; Van Eps, A.W.; Thoefner, M.; Blackall, L.L.; Al Jassim, R.A.M.; Morton, J.M.; Pollitt, C.C. Changes in equine hindgut bacterial populations during oligofructose-induced laminitis. Environ. Microbiol. 2006, 8, 885–898. [Google Scholar] [CrossRef]
- De Fombelle, A.; Julliand, V.; Drogoul, C.; Jacotot, E. Feeding and microbial disorders in horses: 1-effects of an abrupt incorporation of two levels of barley in a hay diet on microbial profile and activities. J. Equine Vet. Sci 2001, 21, 439–445. [Google Scholar] [CrossRef]
- Elzinga, S.; Weese, J.; Adams, A. Comparison of the fecal microbiota in horses with Equine Metabolic Syndrome (EMS) and metabolically normal controls fed a similar all forage diet. J. Equine Vet. Sci 2016, 44, 9–16. [Google Scholar] [CrossRef]
- Dong, H.-j.; Hwang, H.; Han, J.; Cho, S. Diversity of the Gastric Microbiota in Thoroughbred Racehorses Having Gastric Ulcer. J. Microbiol. Biotechnol. 2016, 26, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoster, A.; Mosing, M.; Jalali, M.; Staempfli, H.; Weese, J. Effects of transport, fasting and anaesthesia on the faecal microbiota of healthy adult horses. Equine Vet. J. 2016, 48, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Harlow, B.E.; Lawrence, L.M.; Flythe, M.D.; Hayes, S.H.; Gellin, G.L.; Strasinger, L.A.; Brümmer, M.; Fowler, A.L. Microbial species richness of equine fecal microflora in horses challenged with antibiotics. J. Equine Vet. Sci. 2013, 33, 331. [Google Scholar] [CrossRef]
- Costa, M.; Stampfli, H.; Arroyo, L.; Allen-Vercoe, E.; Gomes, R.; Weese, J. Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Vet. Res. 2015, 11, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, K.A.; Gee, E.K.; Rogers, C.W.; Kittelmann, S.; Biggs, P.J.; Bermingham, E.N.; Bolwell, C.F.; Thomas, D.G. Seasonal variation in the faecal microbiota of mature adult horses maintained on pasture in New Zealand. Animals 2021, 11, 2300. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, K.; Rogers, C.; Gee, E.; Fitch, G.; Bolwell, C.; Kittelmann, S.; Bermingham, E.; Thomas, D.G. Comparison of gastrointestinal transit times in stabled Thoroughbred horses during abrupt dietary transition between freshly cut pasture and three conserved forage-based diets. Anim. Prod. Sci. 2021, accepted. [Google Scholar]
- McGreevy, P.D.; Webster, A.J.F.; Nicol, C.J. Study of the behaviour, digestive efficiency and gut transit times of crib-biting horses. Vet. Rec. 2001, 148, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Henneke, D.R.; Potter, G.D.; Kreider, J.L.; Yeates, B.F. Relationship between condition score, physical measurements and body fat percentage in mares. Equine Vet. J. 1983, 15, 371–372. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Cox, M.; Peterson, D.; Biggs, P. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Gihring, T.M.; Green, S.J.; Schadt, C.W. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ. Microbiol. 2012, 14, 285–290. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.; Ryan, P. PAST: Palaeontological statistics software package for education and data analysis. Plalaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Spellerberg, I.F.; Fedor, P.J. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef] [Green Version]
- Gotelli, N.J.; Colwell, R.K. Estimating species richness. Biol. Divers. Front. Meas. Assess. 2011, 12, 39–54. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [Green Version]
- Folke, C.; Carpenter, S.; Walker, B.; Scheffer, M.; Elmqvist, T.; Gunderson, L.; Holling, C.S. Regime Shifts, Resilience, and Biodiversity in Ecosystem Management. Ann. Rev. Ecol. 2004, 35, 557–581. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.; Holling, C.S.; Carpenter, S.R.; Kinzig, A. Resilience, adaptability and transformability in social—Ecological systems. Ecol. Soc. 2004, 9, 5. [Google Scholar] [CrossRef]
- Costa, M.C.; Silva, G.; Ramos, R.V.; Staempfli, H.R.; Arroyo, L.G.; Kim, P.; Weese, J.S. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Schoster, A.; Arroyo, L.G.; Staempfli, H.R.; Weese, J.S. Comparison of microbial populations in the small intestine, large intestine and feces of healthy horses using terminal restriction fragment length polymorphism. BMC Res. Notes 2013, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Bayer, E.A. Plant Cell Wall Breakdown by Anaerobic Microorganisms from the Mammalian Digestive Tract. Ann. N. Y. Acad. Sci. 2008, 1125, 280–288. [Google Scholar] [CrossRef]
- Moreau, M.M.; Eades, S.C.; Reinemeyer, C.R.; Fugaro, M.N.; Onishi, J.C. Illumina sequencing of the V4 hypervariable region 16S rRNA gene reveals extensive changes in bacterial communities in the cecum following carbohydrate oral infusion and development of early-stage acute laminitis in the horse. Vet. Microbiol. 2014, 168, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.S.; Black, S.J.; Blanchard, J.L. An in vitro model of the horse gut microbiome enables identification of lactate-utilizing bacteria that differentially respond to starch induction. PLoS ONE 2013, 8, e77599. [Google Scholar] [CrossRef] [Green Version]
- Weese, J.S.; Holcombe, S.J.; Embertson, R.M.; Kurtz, K.A.; Roessner, H.A.; Jalali, M.; Wismer, S.E. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 2015, 47, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Leng, J.; Proudman, C.; Blow, F.; Darby, A.; Swann, J. Understanding Intestinal Microbiota in Equine Grass Sickness: Next Generation Sequencing of Faecal Bacterial DNA. Equine Vet. J. 2015, 47, 9. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Stämpfli, H.; Allen-Vercoe, E.; Weese, J. Development of the faecal microbiota in foals. Equine Vet. J. 2015, 48, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.J.; Nielsen, B.D.; Schott, H.C.; Elzinga, S.; Newbold, C.J. Characterisation of the faecal bacterial community in adult and elderly horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biagi, E.; Candela, M.; Fairweather-Tait, S.; Franceschi, C.; Brigidi, P. Ageing of the human metaorganism: The microbial counterpart. Age 2012, 34, 247–267. [Google Scholar] [CrossRef] [Green Version]
- Drogoul, C.; de Fombelle, A.; Julliand, V. Feeding and microbial disorders in horses: 2-effect of three hay, grain ratios on digesta passage rate and digestibility in ponies. J. Equine Vet. Sci 2001, 21, 487–491. [Google Scholar] [CrossRef]
- Bulmer, L.; McBride, S.; Williams, K.; Murray, J.-A. The effects of a high-starch or high-fibre diet on equine reactivity and handling behaviour. Appl. Anim. Behav. Sci. 2015, 165, 95–102. [Google Scholar] [CrossRef]
- Grimm, P.; Julliand, V.; Philippeau, C.; Sadet-Bourgeteau, S. Effect of yeast supplementation on hindgut microbiota and digestibility of horses subjected to an abrupt change of hays. Livest. Sci. 2016, 186, 34–40. [Google Scholar] [CrossRef]
- Julliand, V.; Grimm, P. The microbiome of the horse hindgut: History and current knowledge. J. Anim. Sci. 2016, 94, 2262–2274. [Google Scholar] [CrossRef]
- Blackmore, T.M.; Dugdale, A.; Argo, C.M.; Curtis, G.; Pinloche, E.; Harris, P.A.; Worgan, H.J.; Girdwood, S.E.; Dougal, K.; Newbold, C.J.; et al. Strong Stability and Host Specific Bacterial Community in Faeces of Ponies. PLoS ONE 2013, 8, e75079. [Google Scholar] [CrossRef] [Green Version]
- De Fombelle, A.; Varloud, M.; Goachet, A.G.; Jacotot, E.; Philippeau, C.; Drogoul, C.; Julliand, V. Characterization of the microbial and biochemical profile of the different segments of the digestive tract in horses given two distinct diets. Anim. Sci. 2003, 77, 293–304. [Google Scholar] [CrossRef]
- Clarridge, J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, M.M.; Harris, H.M.B.; Jeffery, I.B.; Claesson, M.J.; Younge, B.; O’Toole, P.W.; Ross, R.P. The core faecal bacterial microbiome of Irish Thoroughbred racehorses. Lett. Appl. Microbiol. 2013, 57, 492–501. [Google Scholar] [CrossRef]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janecka, J.E. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Park, J.-H.; Kang, H.-J.; Lee, Y.H.; Lee, T.J.; Park, H.-D. Distribution and abundance of Spirochaetes in full-scale anaerobic digesters. Bioresour. Technol. 2013, 145, 25–32. [Google Scholar] [CrossRef]
- Frape, D. Equine Nutrition and Feeding; Wiley-Blackwell: Baltimore, MD, USA, 2010. [Google Scholar]
- Cohen, N.; Peloso, J. Risk factors for history of previous colic and for chronic, intermittent colic in a population of horses. J. Am. Vet. Med. Assoc. 1996, 208, 697. [Google Scholar] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, K.; Stewart, C.S.; Flint, H.J.; Shirazi-Beechey, S.P. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol. Ecol. 2001, 38, 141–151. [Google Scholar] [CrossRef]


| Nutrient | Pasture 1 | Pasture 2 | Pasture 3 | FE (Ensiled Lucerne and Timothy) | Hay | Whole Oats |
|---|---|---|---|---|---|---|
| DE MJ/kg | 10.8 ± 0.4 | 11.0 ± 0.4 | 10.7 ± 0.3 | 10.7 ± 0.3 | 9.3 ± 0.3 | 15.1 ± 0.2 |
| % DM | 15.6 ± 1.7 | 16.4 ± 2.5 | 15.8 ± 2.4 | 39.5 ± 1.4 | 80.5 ± 1.3 | 83.4 ± 1.1 |
| % Ash | 13.2 ± 1.0 | 11.8 ± 0.8 | 12.2 ± 1.3 | 7.8 ± 0.5 | 7.6 ± 0.6 | 3.0 ± 0.0 |
| % Crude Protein | 25.0 ± 1.6 | 22.9 ± 1.7 | 19.2 ± 2.7 | 17.3 ± 1.3 | 11.7 ± 0.7 | 10.9 ± 0.2 |
| % Crude Fat | 4.9 ± 0.2 | 4.7 ± 0.4 | 4.4 ± 0.6 | 3.9 ± 0.3 | 1.9 ± 0.4 | 5.9 ± 0.2 |
| % CHO ‡ | 56.9 ± 2.2 | 60.7 ± 2.2 | 64.3 ± 4.1 | 71.0 ± 1.6 | 78.7 ± 1.3 | 80.3 ± 0.3 |
| % ADF | 45.7 ± 2.3 | 46.6 ± 2.4 | 49.3 ± 1.8 | 58.1 ± 3.1 | 62.9 ± 2.2 | 24.4 ± 2.1 |
| % NDF | 26.6 ± 1.6 | 28.2 ± 1.3 | 29.8 ± 1.9 | 39.1 ± 1.3 | 43.7 ± 2.2 | 12.1 ± 1.0 |
| % Lignin | 3.7 ± 0.7 | 4.8 ± 1.1 | 4.9 ± 1.0 | 6.4 ± 0.3 | 7.5 ± 0.6 | 2.5 ± 0.3 |
| % Starch | 1.0 ± 1.0 | 1.4 ± 0.9 | 0.5 ± 0.5 | 2.6 ± 0.6 | 1.3 ± 0.4 | 50.3 ± 3.2 |
| % WSC | 20.6 ± 1.5 | 21.8 ± 2.9 | 23.4 ± 2.4 | 10 ± 1.6 | 9.0 ± 1.2 | NA |
| % ESC | 13.9 ± 2.9 | 13.9 ± 3.9 | 17.7 ± 1.7 | 5.4 ± 1.2 | 6.6 ± 1.3 | NA |
| % NFC § | 30.3 ± 3.0 | 32.5 ± 2.2 | 34.5 ± 4.7 | 31.9 ± 1.6 | 35.0 ± 1.3 | 68.2 ± 1.3 |
| % TDN ϕ | 70.8 ± 2.2 | 70.1 ± 2.4 | 68.3 ± 2.2 | 68.1 ± 1.5 | 59.1 ± 1.0 | 86.6 ± 1.3 |
| RFV | 169 ± 11 | 163 ± 10 | 151 ± 7 | 119 ± 8 | 94 ± 3 | NA |
| Variable | Units | Dietary Treatments | |||||
|---|---|---|---|---|---|---|---|
| Pasture 1 | Pasture 2 | Pasture 3 | FE (Ensiled Lucerne and Timothy) | FE + Whole Oats | Hay + Whole Oats | ||
| Feed offered | kg/d (as-fed basis) | 46 ± 10 | 57 ± 8 | 60 ± 9 | 24 ± 4 | 27 ± 5 | 15 ± 3 |
| Feed consumed | kg/d (as-fed basis) | 38 ± 8 | 44 ± 8 | 46 ± 9 | 19 ± 4 | 19 ± 5 | 9 ± 2 |
| - | kg/d (DM basis) | 6.1 ± 1.3 | 7.0 ± 1.3 | 7.3 ± 1.4 | 7.4 ± 1.5 | 7.6 ± 2.1 | 6.9 ± 1.5 |
| - | g DM/kg BW0.75/d | 54 a ± 11 | 62 b ± 11 | 65 b ± 12 | 66 b ± 12 | 69 b ± 19 | 62 b ± 13 |
| Faeces voided | kg/d (fresh basis) | 10 a ± 3 | 15 b ± 5 | 16 b ± 3 | 15 b ± 5 | 14 bc ± 5 | 12 ac ± 3 |
| Alpha Diversity Index | Diets | p-Value * | |||||
|---|---|---|---|---|---|---|---|
| Pasture 1 | Pasture 2 | Pasture 3 | FE (Ensiled lucerne and Timothy) | FE + Whole Oats | Hay + Whole Oats | - | |
| Simpson’s (diversity) | 0.89 b | 0.89 b | 0.91a | 0.90 ab | 0.89 b | 0.89 b | <0.001 |
| (IQR) | (0.88–0.90) | (0.88–0.90) | (0.90–0.92) | (0.89–0.91) | (0.88–0.90) | (0.88–0.90) | - |
| Shannon-Wiener (entropy) | 2.74 b | 2.75 b | 2.91 a | 2.78 b | 2.80 b | 2.74 b | <0.001 |
| (IQR) | (2.67–2.80) | (2.70–2.80) | (2.87–2.98) | (2.77–2.91) | (2.68–2.88) | (2.64–2.80) | - |
| Chao 1 (richness) | 61 b | 65 ab | 69 a | 62b | 60b | 62b | <0.001 |
| (IQR) | (59–65) | (63–70) | (65–72) | (56–65) | (56–63) | (58–65) | |
| Diets | Phyla | Friedman’s Test | Post-Hoc Analysis | Friedman’s Test | Post-Hoc Analysis |
|---|---|---|---|---|---|
| (Marker Time Points) | (T0 vs. M1) | (Day Time Points) | (T0 vs. T24) | ||
| P2 | Firmicutes | 0.004 ** | 0.046 * | 0.012 ** | 0.077 |
| Bacteroidetes | 0.010 ** | 0.077 | 0.019 * | 0.046 * | |
| Spirochaetes | 0.101 | 0.323 | 0.007 ** | 0.231 | |
| Verrucomicrobia | 0.159 | 0.662 | 0.039 * | 0.446 | |
| Other Phyla | 0.456 | 0.836 | 0.024 * | 0.695 | |
| P3 | Firmicutes | 0.701 | 0.569 | 0.023 * | 0.569 |
| Bacteroidetes | 0.161 | 0.323 | 0.197 | 0.446 | |
| Spirochaetes | 0.012 * | 0.230 | 0.019 * | 0.169 | |
| Verrucomicrobia | 0.180 | 0.077 | 0.004 ** | 0.077 | |
| Other Phyla | 0.491 | 0.001 ** | 0.003 ** | 0.058 | |
| FE | Firmicutes | 0.207 | 0.077 | 0.030 * | 0.077 |
| Bacteroidetes | 0.091 | 0.107 | 0.073 | 0.169 | |
| Spirochaetes | 0.091 | 1.000 | 0.009 ** | 0.446 | |
| Verrucomicrobia | 0.333 | 0.446 | 0.082 | 0.846 | |
| Other Phyla | 0.339 | 0.438 | 0.014 ** | 0.762 | |
| FE + O | Firmicutes | 0.228 | 0.169 | 0.009 ** | 0.046 * |
| Bacteroidetes | 0.029 * | 0.077 | 0.006 ** | 0.046 * | |
| Spirochaetes | 0.077 | 1.000 | 0.048 * | 1.000 | |
| Verrucomicrobia | 0.091 | 0.323 | 0.038 * | 0.692 | |
| Other Phyla | 0.109 | 0.633 | 0.218 | 0.321 | |
| H + O | Firmicutes | 0.334 | 0.846 | 0.066 | 0.846 |
| Bacteroidetes | 0.034 * | 0.446 | 0.006 ** | 0.323 | |
| Spirochaetes | 0.643 | 0.631 | 0.003 ** | 0.569 | |
| Verrucomicrobia | 0.692 | 1.000 | 0.487 | 1.000 | |
| Other Phyla | 0.242 | 0.554 | 0.025 * | 0.194 |
| - | - | Firmicutes | - | Bacteroidetes | - | - | ||
|---|---|---|---|---|---|---|---|---|
| - | Time Point (h) | Mean | SD | % Difference | Mean | SD | % Difference | F:B Ratio |
| Diet P1 | 0 | 0.598 | 0.053 | - | 0.295 | 0.033 | - | 2.03 |
| 24 | 0.522 | 0.129 | −13% | 0.339 | 0.075 | 15% | 1.54 | |
| 48 | 0.653 | 0.076 | 25% | 0.245 | 0.053 | −28% | 2.67 | |
| 72 | 0.516 | 0.028 | −21% | 0.325 | 0.031 | 33% | 1.59 | |
| 96 | 0.557 | 0.070 | 8% | 0.330 | 0.051 | 2% | 1.69 | |
| 120 | 0.520 | 0.049 | −7% | 0.339 | 0.056 | 3% | 1.54 | |
| 144 | 0.616 | 0.099 | 18% | 0.290 | 0.083 | −14% | 2.12 | |
| 168 | 0.534 | 0.045 | −13% | 0.331 | 0.045 | 14% | 1.62 | |
| Diet P2 | 0 | 0.617 | 0.080 | - | 0.245 | 0.069 | - | 2.52 |
| 12 | 0.573 | 0.155 | −7% | 0.209 | 0.112 | −15% | 2.76 | |
| 24 | 0.514 | 0.084 | −10% | 0.361 | 0.050 | 73% | 1.43 | |
| 48 | 0.505 | 0.099 | −2% | 0.375 | 0.073 | 4% | 1.35 | |
| 72 | 0.506 | 0.072 | 0% | 0.364 | 0.072 | −3% | 1.39 | |
| 96 | 0.463 | 0.099 | −8% | 0.403 | 0.071 | 11% | 1.15 | |
| 120 | 0.450 | 0.117 | −3% | 0.412 | 0.104 | 2% | 1.09 | |
| 144 | 0.619 | 0.098 | 38% | 0.280 | 0.086 | −32% | 2.21 | |
| 168 | 0.458 | 0.136 | −26% | 0.406 | 0.096 | 45% | 1.13 | |
| Diet P3 | 0 | 0.553 | 0.094 | - | 0.261 | 0.089 | - | 2.12 |
| 12 | 0.463 | 0.047 | −16% | 0.337 | 0.052 | 29% | 1.38 | |
| 24 | 0.589 | 0.058 | 27% | 0.309 | 0.071 | −8% | 1.91 | |
| 48 | 0.583 | 0.072 | −1% | 0.321 | 0.082 | 4% | 1.82 | |
| 72 | 0.620 | 0.083 | 6% | 0.287 | 0.099 | −11% | 2.17 | |
| 96 | 0.489 | 0.065 | −21% | 0.379 | 0.080 | 32% | 1.29 | |
| 120 | 0.544 | 0.077 | 11% | 0.318 | 0.101 | −16% | 1.71 | |
| 144 | 0.449 | 0.083 | −17% | 0.397 | 0.059 | 25% | 1.13 | |
| 168 | 0.411 | 0.057 | −8% | 0.417 | 0.109 | 5% | 0.99 | |
| Diet FE | 0 | 0.443 | 0.107 | - | 0.415 | 0.099 | - | 1.07 |
| 24 | 0.594 | 0.116 | 34% | 0.264 | 0.103 | −36% | 2.25 | |
| 48 | 0.517 | 0.099 | −13% | 0.332 | 0.062 | 26% | 1.56 | |
| 72 | 0.626 | 0.094 | 21% | 0.251 | 0.062 | −24% | 2.50 | |
| 96 | 0.621 | 0.128 | −1% | 0.212 | 0.108 | −16% | 2.94 | |
| 120 | 0.609 | 0.072 | −2% | 0.249 | 0.063 | 17% | 2.45 | |
| 144 | 0.640 | 0.079 | 5% | 0.246 | 0.081 | −1% | 2.60 | |
| 168 | 0.666 | 0.101 | 4% | 0.190 | 0.094 | −23% | 3.52 | |
| Diet FE + O | 0 | 0.448 | 0.091 | - | 0.412 | 0.072 | - | 1.09 |
| 24 | 0.576 | 0.097 | 29% | 0.296 | 0.080 | −28% | 1.95 | |
| 48 | 0.505 | 0.104 | −12% | 0.319 | 0.074 | 8% | 1.59 | |
| 72 | 0.585 | 0.073 | 16% | 0.281 | 0.063 | −12% | 2.08 | |
| 96 | 0.677 | 0.083 | 16% | 0.195 | 0.068 | −31% | 3.48 | |
| 120 | 0.643 | 0.070 | −5% | 0.237 | 0.067 | 22% | 2.71 | |
| 144 | 0.627 | 0.085 | −2% | 0.227 | 0.077 | −4% | 2.76 | |
| 168 | 0.597 | 0.091 | −5% | 0.255 | 0.089 | 12% | 2.35 | |
| Diet H + O | 0 | 0.513 | 0.099 | - | 0.327 | 0.086 | - | 1.57 |
| 24 | 0.493 | 0.061 | −4% | 0.361 | 0.067 | 10% | 1.37 | |
| 48 | 0.448 | 0.091 | −9% | 0.292 | 0.059 | −19% | 1.54 | |
| 72 | 0.484 | 0.060 | 8% | 0.232 | 0.069 | −21% | 2.09 | |
| 96 | 0.593 | 0.117 | 23% | 0.175 | 0.060 | −25% | 3.40 | |
| 120 | 0.479 | 0.096 | −19% | 0.204 | 0.085 | 17% | 2.35 | |
| 144 | 0.496 | 0.087 | 4% | 0.236 | 0.092 | 16% | 2.10 | |
| 168 | 0.495 | 0.117 | 0% | 0.204 | 0.081 | −14% | 2.43 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, K.A.; Rogers, C.W.; Gee, E.K.; Kittelmann, S.; Bolwell, C.F.; Bermingham, E.N.; Biggs, P.J.; Thomas, D.G. Resilience of Faecal Microbiota in Stabled Thoroughbred Horses Following Abrupt Dietary Transition between Freshly Cut Pasture and Three Forage-Based Diets. Animals 2021, 11, 2611. https://doi.org/10.3390/ani11092611
Fernandes KA, Rogers CW, Gee EK, Kittelmann S, Bolwell CF, Bermingham EN, Biggs PJ, Thomas DG. Resilience of Faecal Microbiota in Stabled Thoroughbred Horses Following Abrupt Dietary Transition between Freshly Cut Pasture and Three Forage-Based Diets. Animals. 2021; 11(9):2611. https://doi.org/10.3390/ani11092611
Chicago/Turabian StyleFernandes, Karlette A., Chris W. Rogers, Erica K. Gee, Sandra Kittelmann, Charlotte F. Bolwell, Emma N. Bermingham, Patrick J. Biggs, and David G. Thomas. 2021. "Resilience of Faecal Microbiota in Stabled Thoroughbred Horses Following Abrupt Dietary Transition between Freshly Cut Pasture and Three Forage-Based Diets" Animals 11, no. 9: 2611. https://doi.org/10.3390/ani11092611






