CRY1 Gene Polymorphism and Racing Performance of Homing Pigeons
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Approval
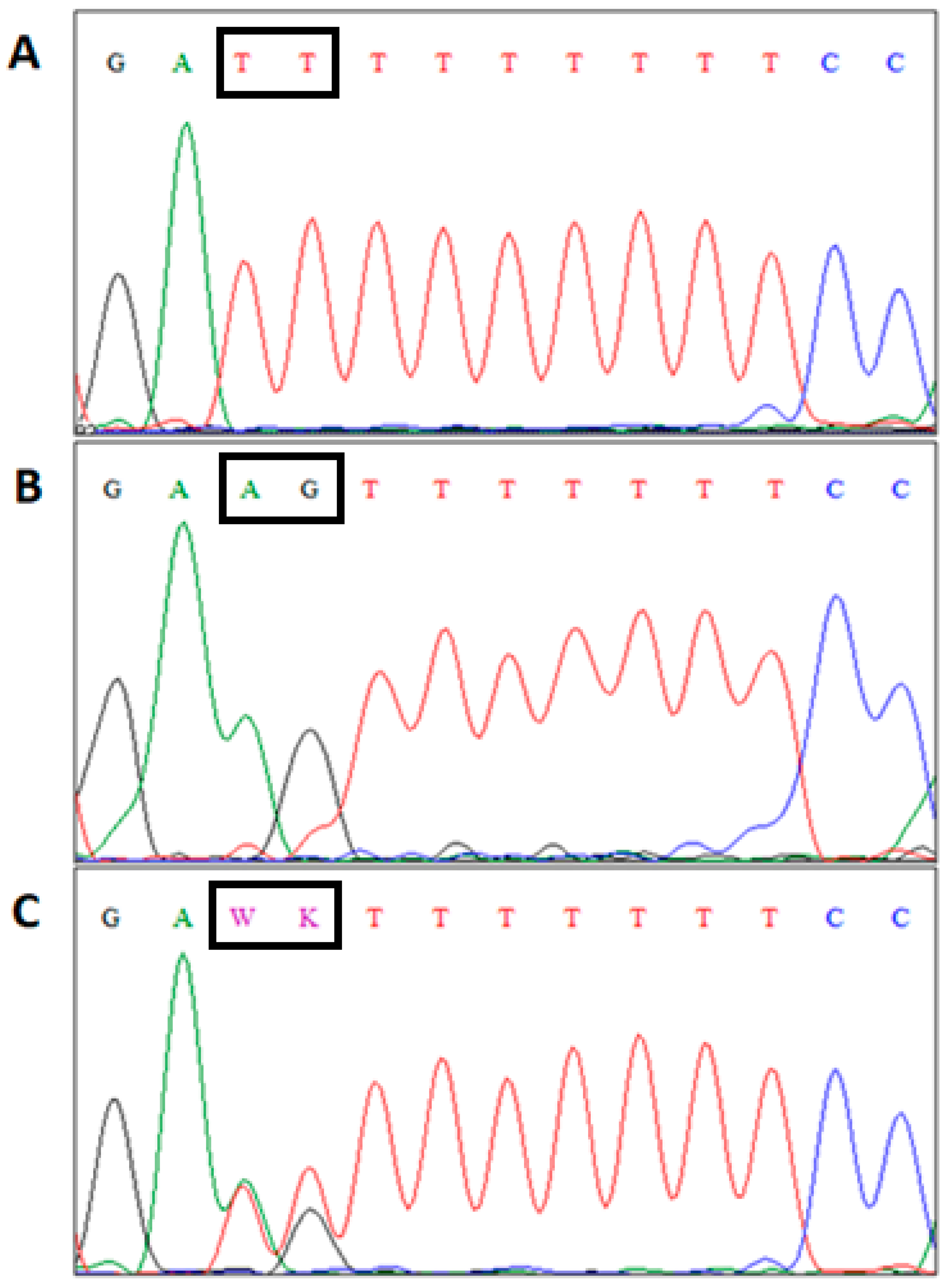
2.2. Animals, DNA Analyses
- CRY1-F: 5′-GGCAACTAACAATCCACGGT-3′
- CRY1-R: 5′-TTGGGCAAAGGGACAGAAAC-3′
- CRY-MluCI-F: 5′-AGAAACATTGGTTACTCTTATAGTTA*A-3′
- CRY-MluCI-R: 5′-AACATTGGGCAAAGGGACAG-3′
2.3. Statistical Analysis
- AP is the ace points value
- a is the number of pigeons on the prize list (20% of participated)
- b is the position of a pigeon in a race
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnsen, S.; Lohmann, K.J. The physics and neurobiology of magnetoreception. Nat. Rev. Neurosci. 2005, 6, 703–712. [Google Scholar] [CrossRef]
- Zhou, Z.; Peng, X.; Chen, J.; Wu, X.; Wang, Y.; Hong, Y. Identification of zebrafish magnetoreceptor and cryptochrome homologs. Sci. China Life Sci. 2016, 59, 1324–1331. [Google Scholar] [CrossRef] [Green Version]
- Hochstoeger, T.; Al Said, T.; Maestre, D.; Walter, F.; Vilceanu, A.; Pedron, M.; Cushion, T.D.; Snider, W.; Nimpf, S.; Nordmann, G.C.; et al. The biophysical, molecular, and anatomical landscape of pigeon CRY4: A candidate light-based quantal magnetosensor. Sci. Adv. 2020, 6, eabb9110. [Google Scholar] [CrossRef] [PubMed]
- Ritz, T.; Adem, S.; Schulten, K. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 2000, 78, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Kirschvink, J.L.; Walker, M.M.; Diebel, C.E. Magnetite-based magnetoreception. Curr. Opin. Neurobiol. 2001, 11, 462–467. [Google Scholar] [CrossRef]
- Nordmann, G.C.; Hochstoeger, T.; Keays, D.A. Magnetoreception-A sense without a receptor. PLoS Biol. 2017, 15, e2003234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.Q.; Dickman, J.D. Magnetoreception in an avian brain in part mediated by inner ear lagena. Curr. Biol. 2011, 21, 418–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkenberg, G.; Fleißner, G.; Schuchart, K.; Kuehbacher, M.; Thalau, P.; Mouritsen, H.; Heyers, D.; Wellenreuther, G.; Fleißner, G. Avian magnetoreception: Elaborate iron mineral containing dendrites in the upper beak seem to be a common feature in birds. PLoS ONE 2010, 5, e9231. [Google Scholar] [CrossRef] [Green Version]
- Schiffner, I.; Wiltschko, R. Temporal fluctuations of the geomagnetic field affect pigeons’ entire homing flight. J. Comp. Physiol. A 2011, 197, 765–772. [Google Scholar] [CrossRef]
- Wiltschko, R.; Wiltschko, W. Sensing magnetic direction in birds: Radical pair processes involving cryptochrome. Biosensors 2014, 4, 221–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.L.; Wang, J.; Pan, W.S.; Liu, Q.J.; Wang, X.J.; Wu, W.J. Observation of magnetic field effects on transient fluorescence spectra of cryptochrome 1 from homing pigeons. Photochem. Photobiol. 2014, 90, 989–996. [Google Scholar] [CrossRef]
- Bolte, P.; Bleibaum, F.; Einwich, A.; Günther, A.; Liedvogel, M.; Heyers, D.; Depping, A.; Wöhlbrand, L.; Rabus, R.; Janssen-Bienhold, U.; et al. Localisation of the Putative Magnetoreceptive Protein Cryptochrome 1b in the Retinae of Migratory Birds and Homing Pigeons. PLoS ONE 2016, 11, e0147819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutta, R.J.; Archipowa, N.; Johannissen, L.O.; Jones, A.R.; Scrutton, N.S. Vertebrate cryptochromes are vestigial flavoproteins. Sci. Rep. 2017, 7, 44906. [Google Scholar] [CrossRef] [Green Version]
- Liedvogel, M.; Maeda, K.; Henbest, K.; Schleicher, E.; Simon, T.; Timmel, C.R.; Hore, P.J.; Mouritsen, H. Chemical magnetoreception: Bird cryptochrome 1a is excited by blue light and forms long-lived radical-pairs. PLoS ONE 2007, 2, e1106. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Chelliah, Y.; Wickramaratne, A.; Jarocha, L.; Karki, N.; Xuc, W.; Mouritsen, H.; Hore, P.J.; Hibbs, R.E.; Green, C.B.; et al. Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon. Proc. Natl. Acad. Sci. USA 2019, 24, 19449–19457. [Google Scholar] [CrossRef] [Green Version]
- Mouritsen, H.; Janssen-Bienhold, U.; Liedvogel, M.; Feenders, G.; Stalleicken, J.; Dirks, P.; Weiler, R. Cryptochromes and neuronal-activity markers colocalize in the retina of migratory birds during magnetic orientation. Proc. Natl. Acad. Sci. USA 2004, 101, 14294–14299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusani, L.; Bertolucci, C.; Frigato, E.; Foà, A. Cryptochrome expression in the eye of migratory birds dependes on their migratory status. J. Exp. Biol. 2014, 217, 918–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinzon-Rodriguez, A.; Bensch, S.; Muheim, R. Expression patterns of cryptochrome genes in avian retina suggest involvement of Cry4 in light-dependent magnetoreception. J. R. Soc. Interface 2018, 15, 20180058. [Google Scholar] [CrossRef] [Green Version]
- Wiltschko, R.; Wiltschko, W. Magnetoreception in birds. J. R. Soc. Interface 2019, 16, 20190295. [Google Scholar] [CrossRef] [Green Version]
- Proskura, W.S.; Cichon, D.; Grzesiak, W.; Zaborski, D.; Sell-Kubiak, E.; Cheng, Y.H.; Dybus, A. Single nucleotide polymorphism in the LDHA gene as a potential marker for the racing performance of pigeons. J. Poult. Sci. 2014, 51, 364–368. [Google Scholar] [CrossRef] [Green Version]
- Proskura, W.S.; Kustosz, J.; Dybus, A.; Lanckriet, R. Polymorphism in dopamine receptor D4 gene is associated with pigeon racing performance. Anim. Genet. 2015, 46, 586–587. [Google Scholar] [CrossRef]
- Proskura, W.S.; Lukaszewicz, A.; Dzierzba, E.; Cichon, D.; Zaborski, D.; Grzesiak, W.; Dybus, A. The Cys83Gly amino acid substitution in feather keratin is associated with pigeon performance in long-distance races. Vet. Med. 2017, 62, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Ramadan, S.; Miyake, T.; Yamaura, J.; Inoue-Murayama, M. LDHA gene is associated with pigeon survivability during racing competitions. PLoS ONE 2018, 13, e0195121. [Google Scholar] [CrossRef] [PubMed]
- Dybus, A.; Yu, Y.H.; Proskura, W.S.; Lanckriet, R. Association of sequence variants in the CKM (Creatine Kinase, M-Type) gene with racing performance of homing pigeons. Russ. J. Genet. 2020, 56, 1006–1011. [Google Scholar] [CrossRef]
- Kim, J.; Williams, F.J.; Dreger, D.L.; Plassais, J.; Davis, B.W.; Parker, H.G.; Ostrander, E.A. Genetic selection of athletic success in sport-hunting dogs. Proc. Natl. Acad. Sci. USA 2018, 115, E7212–E7221. [Google Scholar] [CrossRef] [Green Version]
- Littiere, T.O.; Castro, G.H.F.; Rodriguez, M.d.P.R.; Bonafé, C.M.; Magalhães, A.F.B.; Faleiros, R.R.; Vieira, J.I.G.; Santos, C.G.; Verardo, L.L. Identification and Functional Annotation of Genes Related to Horses’ Performance: From GWAS to Post-GWAS. Animals 2020, 10, 1173. [Google Scholar] [CrossRef]
- Gazda, M.A.; Andrade, P.; Afonso, S.; Dilytė, J.; Archer, J.P.; Lopes, R.J.; Faria, R.; Carneiro, M. Signatures of Selection on Standing Genetic Variation Underlie Athletic and Navigational Performance in Racing Pigeons. Mol. Biol. Evol. 2018, 35, 1176–1189. [Google Scholar] [CrossRef]
- Shao, Y.; Tian, H.-Y.; Hang, J.-J.; Kharrati-Koopaee, H.; Guo, X.; Zhuang, X.-L.; Li, M.-L.; Nanaie, H.A.; DehghaniTafti, E.; Shojaei, B.; et al. Genomic and Phenotypic Analyses Reveal Mechanisms Underlying Homing Ability in Pigeon. Mol. Biol. Evol. 2020, 37, 134–148. [Google Scholar] [CrossRef]
- Jerolmack, C. Animal archaeology: Domestic pigeons and the nature-culture dialectic. Qual. Sociol. Rev. 2007, 3, 74–95. [Google Scholar]
- Biro, D. Homing pigeons. Curr. Biol. 2018, 28, R966–R967. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, K.J.; Cain, S.D.; Dodge, S.A.; Lohmann, C.M.F. Regional magnetic fields as navigational markers for sea turtles. Science 2001, 294, 364–366. [Google Scholar] [CrossRef] [Green Version]
- Chernetsov, N.; Pakhomov, A.; Kobylkov, D.; Kishkinev, D.; Holland, R.A.; Mouritsen, H. Migratory Eurasian reed warblers can use magnetic declination to solve the longitude problem. Curr. Biol. 2017, 27, 2647–2651.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, T.E.; Rayner, M.J.; Walker, M.M. Evidence that pigeons orient to geomagnetic intensity during homing. Proc. R. Soc. Lond. B 2007, 274, 1153–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, K.; Robinson, A.J.; Henbest, K.B.; Hogben, H.J.; Biskup, T.; Ahmad, M.; Schleicher, E.; Weber, S.; Timmel, C.R.; Hore, P.J. Magnetically sensitive light-induced reactions in cryptochrome are consistent with its proposed role as a magnetoreceptor. Proc. Natl. Acad. Sci. USA 2012, 109, 4774–4779. [Google Scholar] [CrossRef] [Green Version]
- Hore, P.J.; Mouritsen, H. The radical-pair mechanism of magnetoreception. Annu. Rev. Biophys. 2016, 45, 299–344. [Google Scholar] [CrossRef]
- Coolidge, C.J.; Seely, R.J.; Patton, J.G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997, 25, 888–896. [Google Scholar] [CrossRef] [Green Version]
- Gallegos, J.E.; Rose, A.B. An intron-derived motif strongly increases gene expression from transcribed sequences through a splicing independent mechanism in Arabidopsis thaliana. Sci. Rep. 2019, 9, 13777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuenca-Bono, B.; García-Molinero, V.; Pascual-García, P.; Dopazo, H.; Llopis, A.; Vilardell, J.; Rodríguez-Navarro, S. SUS1 introns are required for efficient mRNA nuclear export in yeast. Nucleic Acids Res. 2011, 39, 8599–8611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-F.; Feng, L.; Niu, D.-K. Relationship between mRNA stability and intron presence. Biochem. Biophys. Res. Commun. 2007, 354, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Akua, T.; Shaul, O. The Arabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5′ UTR intron. J. Exp. Bot. 2013, 64, 4255–4270. [Google Scholar] [CrossRef] [Green Version]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Förch, P.; Puig, O.; Martínez, C.; Séraphin, B.; Valcárcel, J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5’ splice sites. EMBO J. 2002, 21, 6882–6892. [Google Scholar] [CrossRef]
- Izquierdo, J.M.; Majós, N.; Bonnal, S.; Martínez, C.; Castelo, R.; Guigó, R.; Bilbao, D.; Valcárcel, J. Regulation of Fas Alternative Splicing by Antagonistic Effects of TIA-1 and PTB on Exon Definition. Mol. Cell 2005, 19, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lin, K.-T.; Yang, Y.; Bai, J.; Wang, L.; Sun, J.; Sheng, L.; Krainer, A.R.; Hua, Y. Systematic characterization of short intronic splicing-regulatory elements. bioRxiv 2021. [Google Scholar] [CrossRef]
- Roca, X.; Krainer, A.R.; Eperon, I.C. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013, 27, 129–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramowicz, A.; Gos, M. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Hua, P.; Liu, W.; Chen, D.; Zhao, Y.; Chen, L.; Zhang, N.; Wang, C.; Guo, S.; Wang, L.; Xiao, H.; et al. Cry1 and Tef gene polymorphisms are associated with Major Depressive Disorder in the Chinese population. J. Affect. Disord. 2014, 157, 100–103. [Google Scholar] [CrossRef] [Green Version]
- Onat, O.E.; Kars, M.E.; Gül, Ş.; Bilguvar, K.; Wu, Y.; Özhan, A.; Aydın, C.; Başak, A.N.; Russo, M.A.; Goracci, A.; et al. Human CRY1 variants associate with attention deficit/hyperactivity disorder. J. Clin. Investig. 2020, 130, 3885–3900. [Google Scholar] [CrossRef]
| Genotypes | Alleles | HWE | ||||
|---|---|---|---|---|---|---|
| AG/AG | AG/TT | TT/TT | AG | TT | Χ2 | p |
| 0.87 (n = 107) | 0.11 (n = 14) | 0.02 (n = 2) | 0.93 | 0.07 | 3.1811 | 0.9268 |
| Genotype | All Races | Short Races (100–400 km) | Long Races (500–800 km) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RR | AP | SE | RR | AP | SE | RR | AP | SE | |
| AG/AG | 1017 | 25.61 | 1.07 | 666 | 27.01 * | 1.36 | 351 | 22.94 | 1.73 |
| AG/TT | 176 | 33.79 | 2.86 | 118 | 34.02 * | 3.55 | 58 | 33.31 | 4.89 |
| TT/TT | 16 | 28.16 | 9.45 | 12 | 30.90 | 11.70 | 4 | 19.94 | 15.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dybus, A.; Kulig, H.; Yu, Y.-H.; Lanckriet, R.; Proskura, W.; Cheng, Y.-H. CRY1 Gene Polymorphism and Racing Performance of Homing Pigeons. Animals 2021, 11, 2632. https://doi.org/10.3390/ani11092632
Dybus A, Kulig H, Yu Y-H, Lanckriet R, Proskura W, Cheng Y-H. CRY1 Gene Polymorphism and Racing Performance of Homing Pigeons. Animals. 2021; 11(9):2632. https://doi.org/10.3390/ani11092632
Chicago/Turabian StyleDybus, Andrzej, Hanna Kulig, Yu-Hsiang Yu, Ruben Lanckriet, Witold Proskura, and Yeong-Hsiang Cheng. 2021. "CRY1 Gene Polymorphism and Racing Performance of Homing Pigeons" Animals 11, no. 9: 2632. https://doi.org/10.3390/ani11092632
APA StyleDybus, A., Kulig, H., Yu, Y.-H., Lanckriet, R., Proskura, W., & Cheng, Y.-H. (2021). CRY1 Gene Polymorphism and Racing Performance of Homing Pigeons. Animals, 11(9), 2632. https://doi.org/10.3390/ani11092632






