Domestication Explains Two-Thirds of Differential-Gene-Expression Variance between Domestic and Wild Animals; The Remaining One-Third Reflects Intraspecific and Interspecific Variation
Abstract
:Simple Summary
Abstract
1. Introduction
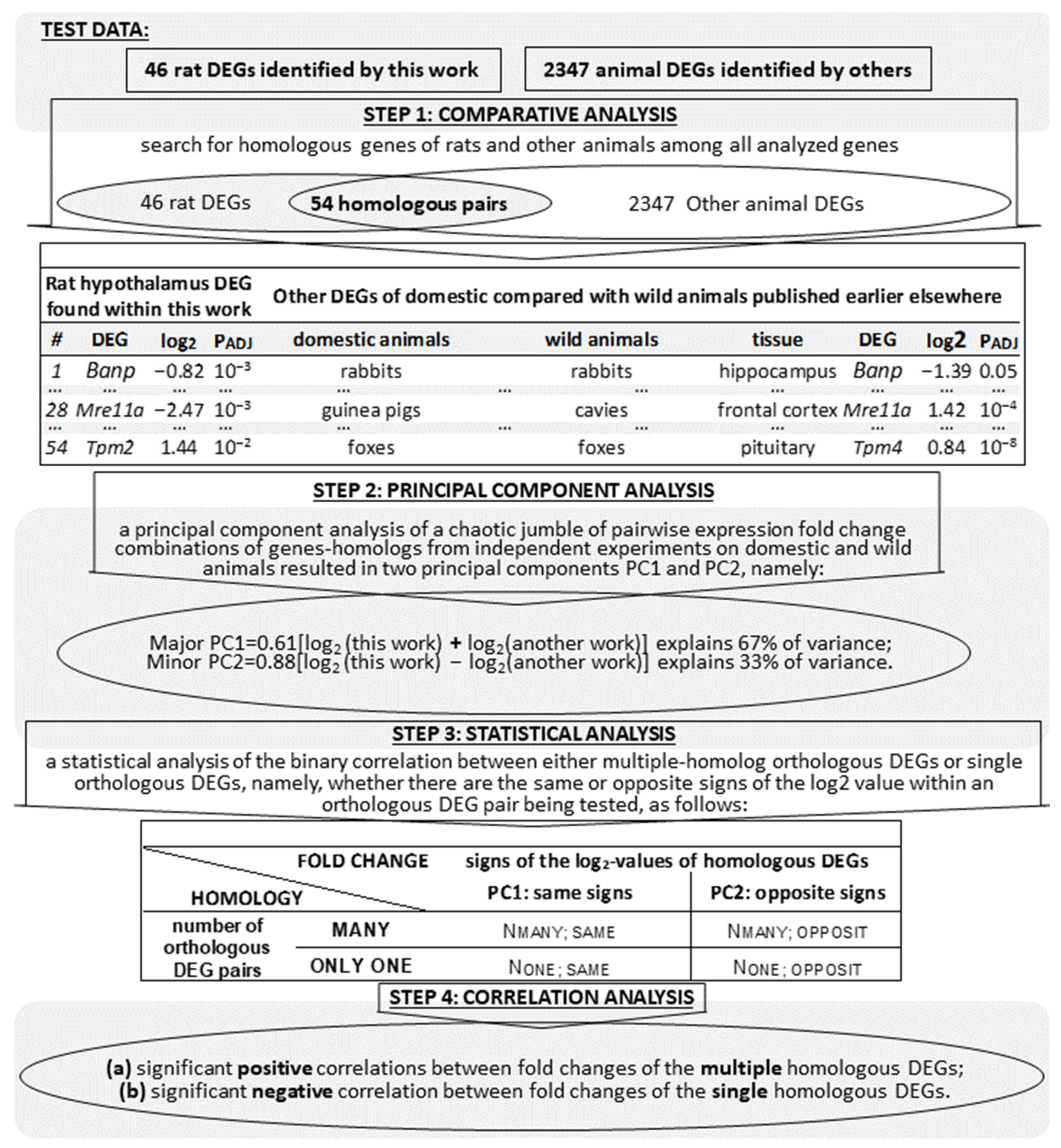
2. Materials and Methods
2.1. Animals
2.2. RNA-Seq
2.3. Mapping of RNA Sequences to the R. norvegicus Reference Genome
2.4. qPCR
2.5. DEGs of Domestic Animals versus Their Wild Congeners
2.6. Statistical Analysis
3. Results
3.1. RNA-Seq and Mapping to the Reference Rat Genome
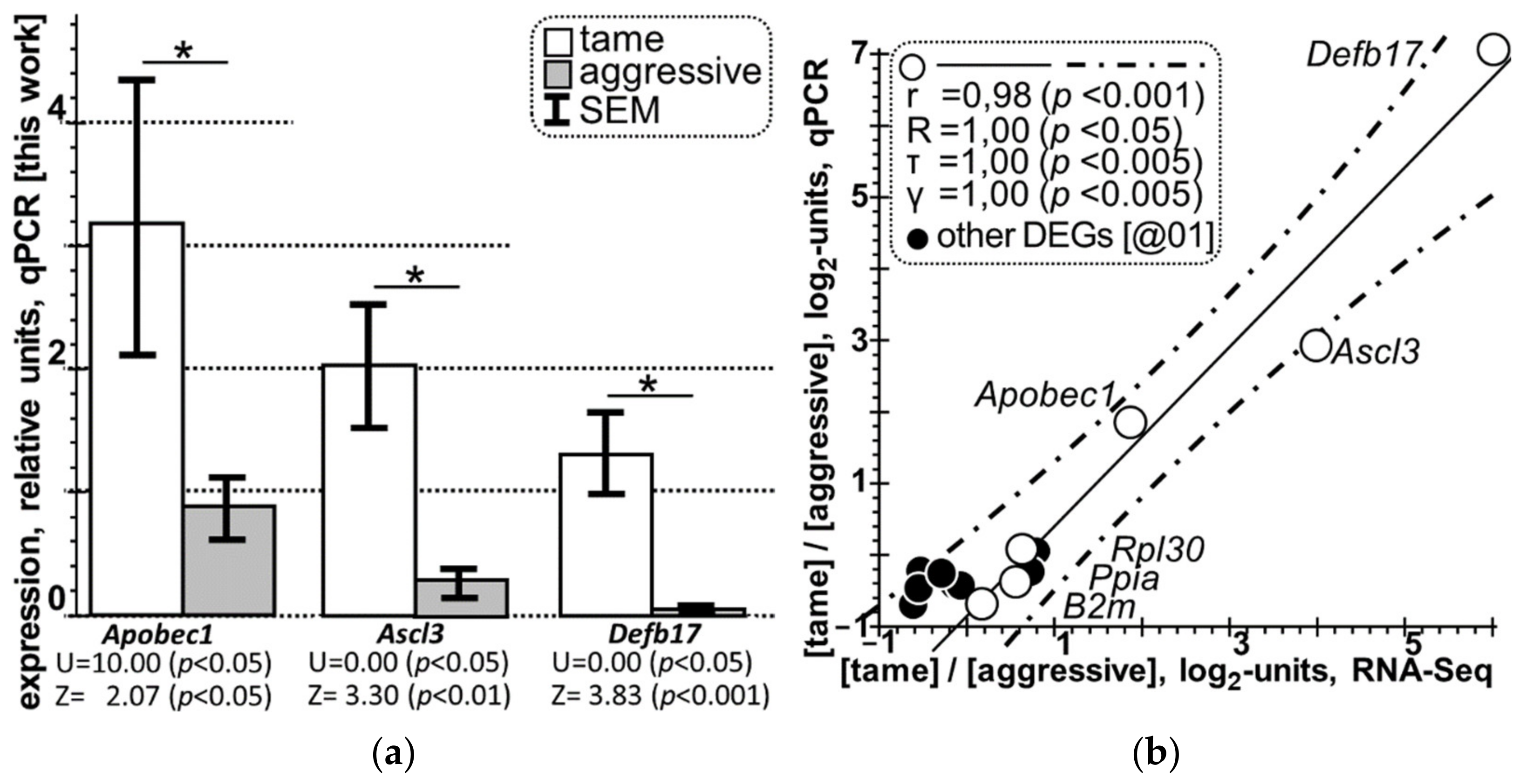
3.2. qPCR Selective Verification of the DEGs Identified in this Work in the Hypothalamus of Tame versus Aggressive Rats
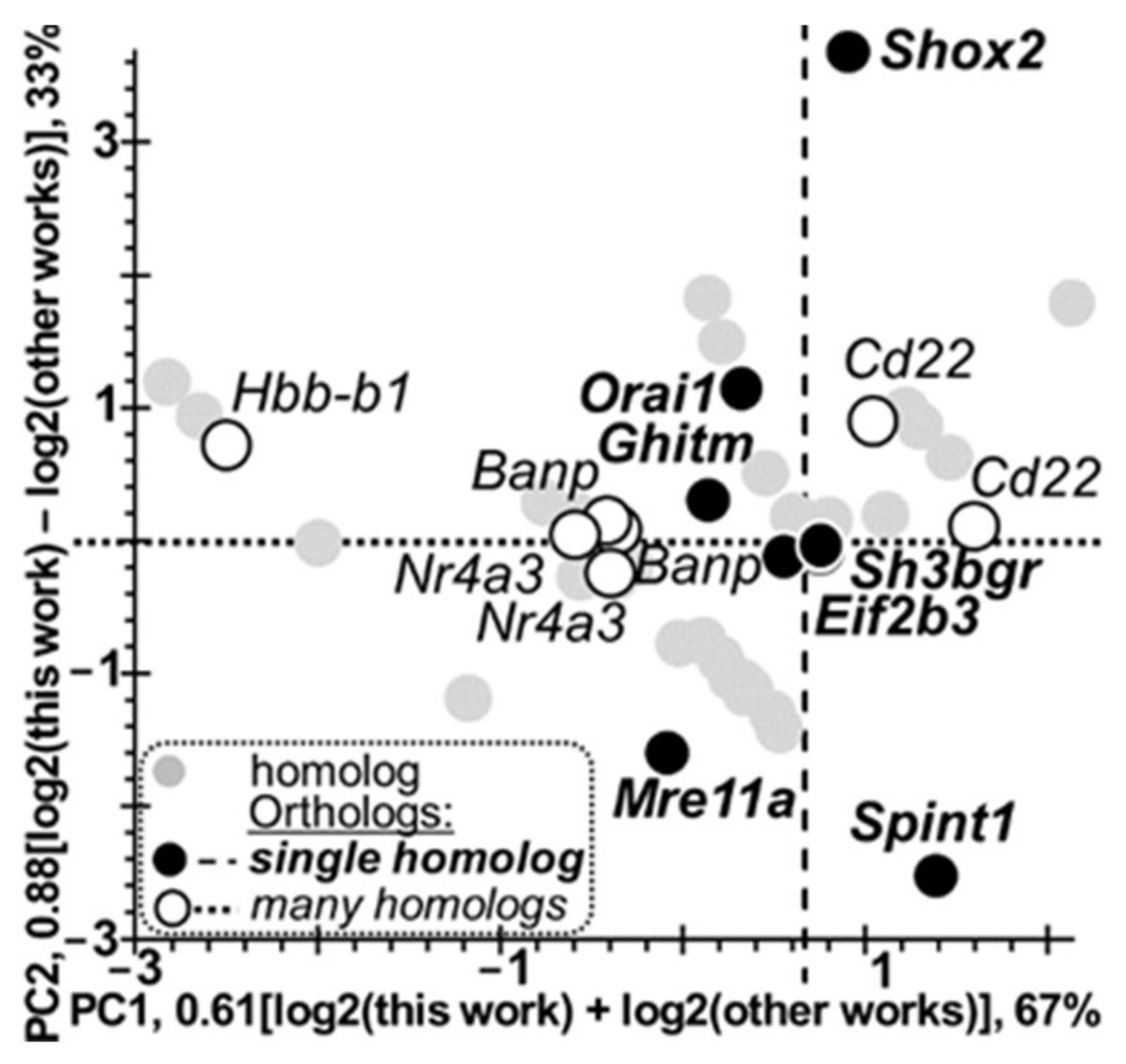
3.3. Verification of the DEGs Found Here in the Hypothalamus of Tame versus Aggressive Rats with Respect to Their Known Homologous DEGs in Domestic and Wild Animals (All Data That We Could Find)
4. Discussion
| Humans | Animals | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Effect of Gene Expression Changes on Human Reproductive Potential, Namely ($): Decreased (→) or Increased (←) | RNA-Seq | Effect of Gene Expression Changes during DIVERGENCE from the most Recent Common Ancestor | [Ref] | |||||
| Deficit (↓) | $ | Excess (↑) | $ | DEG | log2 | Deficit (↓) | Excess (↑) | ||
| i | ii | iii | iv | v | vi | vii | viii | ix | x |
| HBD | hemoglobin deficit (thalassemia) elevates risks of auto-aggressive impulsiveness up to suicide [64], female subfertility [65], causes under-threshold IQ and severe anxiety in children [66] | → | in cohort studies: elite athletes do high-altitude training rising hemoglobin level before low-altitude matches thereby increasing their chances of winning [76] | ← | Hbb-b1 | −3.97 | tame rat | aggressive rat | [75] |
| Hbbl | −5.92 | dogs | wolves | [24] | |||||
| Hba1 | −4.06 | dogs | wolves | [24] | |||||
| Hbad | −1.07 | domestic chicken | wild chicken | [29] | |||||
| Hbm | −6.46 | dogs | wolves | [24] | |||||
| Hbz1 | −7.10 | dogs | wolves | [24] | |||||
| NR5A1 | within human disease models based on Nr5a1-null male mice: hyper-anxiety during impaired aggressive sexual behavior up to male infertility in line with male patients carrying NR5A1 defects [77] as well as NR5A1 deficit can cause hypoestrogenism [78] leading to 1% of cases of female infertility [79] | → | in retrospective meta-analysis of PubMed content: NR5A1 excess contributes to excessive estrogen biosynthesis raising risks of estrogen-dependent inflammatory disorders in women [80] and vice versa for men [81] | → | Nr4a3 | −1,29 | tame rat | aggressive rat | [82] |
| Nr4a3 | −0.85 | domestic chicken | wild chicken | [29] | |||||
| Nr4a3 | −1.58 | domestic rabbits | wild rabbits | [28] | |||||
| Nr2c1 | −0.74 | guinea pigs | cavy | [25] | |||||
| Nr3c1 | 0.51 | wild chicken | domestic chicken | [29] | |||||
| Nr5a1 | −2.19 | guinea pigs | cavy | [25] | |||||
| SHOX | in cohort studies: low SHOX expression causes short stature [83] as adaptive epigenetic response to adverse living conditions, when each calorie saved due to short stature helps to enhance stress resistance [84] | ← | in cohort studies: girls carrying one extra SHOX copy have tall stature [85] elevating risks of pregnancy complications in military active-duty women [86] | → | Shox2 | 6.18 | aggressive rat | tame rat | [87] |
| Shox2 | −3.43 | domestic rabbits | wild rabbits | [28] | |||||
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Belyaev, D.K. Destabilizing selection as a factor in domestication. J. Hered. 1979, 70, 301–308. [Google Scholar] [CrossRef]
- Belyaev, D.K.; Evsikov, V.; Matysko, E.K. Genetics of animal fertility. 3. Effect of monohybrid heterosis on fertility and viability of minks, and prospects of its use in breeding. Sov. Genet. 1972, 8, 46–51. [Google Scholar]
- Belyaev, D.K.; Trut, L.N.; Ruvinsky, A.O. Genetics of the W Locus in Foxes and Expression of its Lethal Effects. J. Hered. 1975, 66, 331–338. [Google Scholar] [CrossRef]
- Belyaev, D.K.; Gruntenko, E.V. Strain Differences in Thymus Weight in Mice with Different Predispositions to Spontaneous Mammary Cancer. Nature 1972, 237, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Osadchuk, A.V.; Markel’, A.L.; Khusainov, R.; Naumenko, E.V.; Beliaev, D.K. Problems in the genetics of stress. IV. A genetic analysis of the level of autonomic reactivity in emotional stress in rats. Genetika 1979, 15, 1847–1857. [Google Scholar] [PubMed]
- Ponomarenko, M.; Gunbin, K.; Doroshkov, A.; Kolchanov, N. Cladogenesis. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; Volume 2, pp. 21–24. [Google Scholar]
- Bray, E.E.; Otto, C.M.; Udell, M.A.R.; Hall, N.J.; Johnston, A.M.; MacLean, E.L. Enhancing the selection and performance of working dogs. Front. Vet. Sci. 2021, 8, 644431. [Google Scholar] [CrossRef]
- Eason, C.T.; Adams, S.L.; Puddick, J.; Romanazzi, D.; Miller, M.R.; King, N.; Johns, S.; Forbes-Blom, E.; Hessian, P.A.; Stamp, L.K.; et al. Greenshell™ Mussels: A Review of Veterinary Trials and Future Research Directions. Vet. Sci. 2018, 5, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Tang, Y.; Huang, Y. Gut health: The results of microbial and mucosal immune interactions in pigs. Anim. Nutr. 2021, 7, 282–294. [Google Scholar] [CrossRef]
- Ma, B.Y.; Gong, Q.L.; Sheng, C.Y.; Liu, Y.; Ge, G.Y.; Li, D.L.; Diao, N.C.; Shi, K.; Li, J.M.; Sun, Z.B.; et al. Prevalence of bovine leukemia in 1983-2019 in China: A systematic review and meta-analysis. Microb. Pathog. 2021, 150, 104681. [Google Scholar] [CrossRef]
- Orlando, L. The Evolutionary and Historical Foundation of the Modern Horse: Lessons from Ancient Genomics. Annu. Rev. Genet. 2020, 54, 563–581. [Google Scholar] [CrossRef]
- Thorne, J.W.; Murdoch, B.M.; Freking, B.A.; Redden, R.R.; Murphy, T.W.; Taylor, J.B.; Blackburn, H.D. Evolution of the sheep industry and genetic research in the United States: Opportunities for convergence in the twenty-first century. Anim. Genet. 2021, 52, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Pathak, P.; Nayak, V.K.; Sinha, R.; Ganaie, B.A. Review on small ruminant conservation status and prospects in India. Trop. Anim. Health Prod. 2020, 52, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, U.; Rao, S.V.R.; Raju, M.V.L.N.; Chatterjee, R.N. Backyard poultry farming for sustained production and enhanced nutritional and livelihood security with special reference to India: A review. Trop. Anim. Health Prod. 2021, 53, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Han, C.; Deng, D.; Ye, F.; Gan, X.; Liu, H.; Li, L.; Xu, H.; Wei, S. Research progress into the physiological changes in metabolic pathways in waterfowl with hepatic steatosis. Br. Poult. Sci. 2020, 62, 118–124. [Google Scholar] [CrossRef]
- Gao, G.; Gao, D.; Zhao, X.; Xu, S.; Zhang, K.; Wu, R.; Yin, C.; Li, J.; Xie, Y.; Hu, S.; et al. Genome-Wide Association Study-Based Identification of SNPs and Haplotypes Associated with Goose Reproductive Performance and Egg Quality. Front. Genet. 2021, 12, 602583. [Google Scholar] [CrossRef]
- Courtier-Orgogozo, V.; Martin, A. The coding loci of evolution and domestication: Current knowledge and implications for bio-inspired genome editing. J. Exp. Biol. 2020, 223, jeb208934. [Google Scholar] [CrossRef] [Green Version]
- Cowan, M.A.; Callan, M.N.; Watson, M.J.; Watson, D.M.; Doherty, T.S.; Michael, D.R.; Dunlop, J.A.; Turner, J.M.; Moore, H.A.; Watchorn, D.J.; et al. Artificial refuges for wildlife conservation: What is the state of the science? Biol. Rev. 2021. [Google Scholar] [CrossRef]
- Belyaev, D.K.; Borodin, P.M. The influence of stress on variation and its role in evolution. Biol. Zent. 1982, 100, 705–714. [Google Scholar]
- Plyusnina, I.; Oskina, I. Behavioral and Adrenocortical Responses to Open-Field Test in Rats Selected for Reduced Aggressiveness Toward Humans. Physiol. Behav. 1997, 61, 381–385. [Google Scholar] [CrossRef]
- McGonigle, P.; Ruggeri, B. Animal models of human disease: Challenges in enabling translation. Biochem. Pharmacol. 2014, 87, 162–171. [Google Scholar] [CrossRef]
- Lutz, C.; Maher, L.; Lee, C.; Kang, W. COVID-19 preclinical models: Human angiotensin-converting enzyme 2 transgenic mice. Hum. Genom. 2020, 14, 1–11. [Google Scholar] [CrossRef]
- Hekman, J.P.; Johnson, J.; Edwards, W.; Vladimirova, A.V.; Gulevich, R.G.; Ford, A.L.; Kharlamova, A.V.; Herbeck, Y.; Acland, G.M.; Raetzman, L.T.; et al. Anterior Pituitary Transcriptome Suggests Differences in ACTH Release in Tame and Aggressive Foxes. G3 Genes Genomes Genet. 2018, 8, 859–873. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhang, H.; Shang, J.; Liu, G.; Xia, T.; Zhao, C.; Sun, G.; Dou, H. Comparative analysis of the blood transcriptomes between wolves and dogs. Anim. Genet. 2018, 49, 291–302. [Google Scholar] [CrossRef]
- Albert, F.W.; Somel, M.; Carneiro, M.; Aximu-Petri, A.; Halbwax, M.; Thalmann, O.; Blanco-Aguiar, J.; Plyusnina, I.Z.; Trut, L.; Villafuerte, R.; et al. A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals. PLoS Genet. 2012, 8, e1002962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, K.; Mao, K.; Che, T.; Zhang, J.; Qiu, W.; Wang, Y.; Tang, Q.; Ma, J.; Li, M.; Li, X. Transcriptome differences in frontal cortex between wild boar and domesticated pig. Anim. Sci. J. 2018, 89, 848–857. [Google Scholar] [CrossRef]
- Yang, Y.; Adeola, A.C.; Xie, H.B.; Zhang, Y.P. Genomic and transcriptomic analyses reveal selection of genes for puberty in Bama Xiang pigs. Zool. Res. 2018, 39, 424–430. [Google Scholar] [CrossRef]
- Sato, D.X.; Rafati, N.; Ring, H.; Younis, S.; Feng, C.; Blanco-Aguiar, J.A.; Rubin, C.-J.; Villafuerte, R.; Hallböök, F.; Carneiro, M.; et al. Brain Transcriptomics of Wild and Domestic Rabbits Suggests That Changes in Dopamine Signaling and Ciliary Function Contributed to Evolution of Tameness. Genome Biol. Evol. 2020, 12, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Fallahshahroudi, A.; Lotvedt, P.; Belteky, J.; Altimiras, J.; Jensen, P. Changes in pituitary gene expression may underlie multiple domesticated traits in chickens. Heredity 2019, 122, 195–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theofanopoulou, C.; Gastaldon, S.; O’Rourke, T.; Samuels, B.D.; Messner, A.; Martins, P.T.; Delogu, F.; Alamri, S.; Boeckx, C. Self-domestication in Homo sapiens: Insights from comparative genomics. PLoS ONE 2017, 12, e0185306. [Google Scholar] [CrossRef] [Green Version]
- Del Savio, L.; Mameli, M. Human domestication and the roles of human agency in human evolution. Hist. Philos. Life Sci. 2020, 42, 21. [Google Scholar] [CrossRef]
- Gunbin, K.; Ponomarenko, M.P.; Suslov, V.V.; Gusev, F.; Fedonin, G.G.; Rogaev, E.I. Evolution of Brain Active Gene Promoters in Human Lineage Towards the Increased Plasticity of Gene Regulation. Mol. Neurobiol. 2017, 55, 1871–1904. [Google Scholar] [CrossRef]
- Chadaeva, I.; Ponomarenko, P.; Rasskazov, D.; Sharypova, E.; Kashina, E.; Kleshchev, M.; Ponomarenko, M.; Naumenko, V.; Savinkova, L.; Kolchanov, N.; et al. Natural Selection Equally Supports the Human Tendencies in Subordination and Domination: A Genome-Wide Study With in silico Confirmation and in vivo Validation in Mice. Front. Genet. 2019, 10, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshchepkov, D.; Ponomarenko, M.; Klimova, N.; Chadaeva, I.; Bragin, A.; Sharypova, E.; Shikhevich, S.; Kozhemyakina, R. A Rat Model of Human Behavior Provides Evidence of Natural Selection against Underexpression of Aggressiveness-Related Genes in Humans. Front. Genet. 2019, 10, 1267. [Google Scholar] [CrossRef] [Green Version]
- Tenenhaus, M.; Tenenhaus, A.; Groenen, P.J.F. Regularized Generalized Canonical Correlation Analysis: A Framework for Sequential Multiblock Component Methods. Psychometrika 2017, 82, 737–777. [Google Scholar] [CrossRef]
- Klimova, N.V.; Chadaeva, I.V.; Shichevich, S.G.; Kozhemyakina, R.V. Differential expression of 10 genes in the hypothalamus of two generations of rats selected for a reaction to humans. Vavilovskii Zhurnal Genet. Sel. 2021, 25, 208–215. [Google Scholar]
- Naumenko, E.; Popova, N.; Nikulina, E.; Dygalo, N.; Shishkina, G.; Borodin, P.; Markel, A. Behavior, adrenocortical activity, and brain monoamines in Norway rats selected for reduced aggressiveness towards man. Pharmacol. Biochem. Behav. 1989, 33, 85–91. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C.R. The Rat Brain in Stereotaxic Coordinates, 7th ed.; Academic Press: London, UK, 2013; p. 472. [Google Scholar]
- Walker, S.E.; Papilloud, A.; Huzard, D.; Sandi, C. The link between aberrant hypothalamic–pituitary–adrenal axis activity during development and the emergence of aggression—Animal studies. Neurosci. Biobehav. Rev. 2018, 91, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Beck, J.; Bolton, E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 49, D10–D17. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Chen, Y.; Wang, D.W.; Liu, X.H. Validation of reference genes via qRT-PCR in multiple conditions in brandt’s voles, lasiopodomys brandtii. Animals 2021, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Zamani, A.; Powell, K.L.; May, A.; Semple, B.D. Validation of reference genes for gene expression analysis following experimental traumatic brain injury in a pediatric mouse model. Brain Res. Bull. 2020, 156, 43–49. [Google Scholar] [CrossRef]
- Gholami, K.; Loh, S.Y.; Salleh, N.; Lam, S.K.; Hoe, S.Z. Selection of suitable endogenous reference genes for qPCR in kidney and hypothalamus of rats under testosterone influence. PLoS ONE 2017, 12, e0176368. [Google Scholar] [CrossRef] [Green Version]
- Penning, L.C.; Vrieling, H.E.; Brinkhof, B.; Riemers, F.M.; Rothuizen, J.; Rutteman, G.R.; Hazewinkel, H.A. A validation of 10 feline reference genes for gene expression measurements in snap-frozen tissues. Vet. Immunol. Immunopathol. 2007, 120, 212–222. [Google Scholar] [CrossRef]
- Lu, Z. PubMed and beyond: A survey of web tools for searching biomedical literature. Database 2011, 2011, baq036. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.N. Animal ecology, with special reference to insects. J. Nerv. Ment. Dis. 1933, 78, 680. [Google Scholar] [CrossRef]
- Pianka, E.R. Natural Selection of Optimal Reproductive Tactics. Am. Zool. 1976, 16, 775–784. [Google Scholar] [CrossRef]
- Chadaeva, I.; Ponomarenko, P.M.; Rasskazov, D.A.; Sharypova, E.B.; Kashina, E.V.; Zhechev, D.A.; Drachkova, I.A.; Arkova, O.V.; Savinkova, L.K.; Ponomarenko, M.P.; et al. Candidate SNP markers of reproductive potential are predicted by a significant change in the affinity of TATA-binding protein for human gene promoters. BMC Genom. 2018, 19. [Google Scholar] [CrossRef] [Green Version]
- Ponomarenko, M.; Kleshchev, M.; Ponomarenko, P.; Chadaeva, I.; Sharypova, E.; Rasskazov, D.; Kolmykov, S.; Drachkova, I.; Vasiliev, G.; Gutorova, N.; et al. Disruptive natural selection by male reproductive potential prevents underexpression of protein-coding genes on the human Y chromosome as a self-domestication syndrome. BMC Genet. 2020, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Vasiliev, G.; Chadaeva, I.; Rasskazov, D.; Ponomarenko, P.; Sharypova, E.; Drachkova, I.; Bogomolov, A.; Savinkova, L.; Ponomarenko, M.; Kolchanov, N.; et al. A Bioinformatics Model of Human Diseases on the Basis of Differentially Expressed Genes (of Domestic Versus Wild Animals) That Are Orthologs of Human Genes Associated with Reproductive-Potential Changes. Int. J. Mol. Sci. 2021, 22, 2346. [Google Scholar] [CrossRef]
- Klimova, N.V.; Oshchepkova, E.; Chadaeva, I.; Sharypova, E.; Ponomarenko, P.; Drachkova, I.; Rasskazov, D.; Oshchepkov, D.; Ponomarenko, M.; Savinkova, L.; et al. Disruptive Selection of Human Immunostimulatory and Immunosuppressive Genes Both Provokes and Prevents Rheumatoid Arthritis, Respectively, as a Self-Domestication Syndrome. Front. Genet. 2021, 12, 610774. [Google Scholar] [CrossRef] [PubMed]
- Samet, H. A top-down quadtree traversal algorithm. IEEE Trans. Pattern Anal. Mach. Intell. 1985, 7, 94–98. [Google Scholar] [CrossRef]
- Sun, G.-L.; Shen, W.; Wen, J.-F. Triosephosphate Isomerase Genes in Two Trophic Modes of Euglenoids (Euglenophyceae) and Their Phylogenetic Analysis. J. Eukaryot. Microbiol. 2008, 55, 170–177. [Google Scholar] [CrossRef]
- Morozova, O.V.; Alekseeva, A.E.; Sashina, T.A.; Brusnigina, N.F.; Epifanova, N.V.; Kashnikov, A.U.; Zverev, V.V.; Novikova, N.A. Phylodynamics of G4P[8] and G2P[4] strains of rotavirus A isolated in Russia in 2017 based on full-genome analyses. Virus Genes 2020, 56, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Hakizimana, J.N.; Yona, C.; Kamana, O.; Nauwynck, H.; Misinzo, G. African Swine Fever Virus Circulation between Tanzania and Neighboring Countries: A Systematic Review and Meta-Analysis. Viruses 2021, 13, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Katoh, T.K.; Finet, C.; Izumitani, H.F.; Toda, M.J.; Watabe, H.-A.; Katoh, T. Phylogeny and evolution of mycophagy in the Zygothrica genus group (Diptera: Drosophilidae). Mol. Phylogenet. Evol. 2021, 163, 107257. [Google Scholar] [CrossRef] [PubMed]
- Namazi, M.R. Minor thalassemia may be a risk factor for impulsiveness. Med. Hypotheses 2003, 60, 335–336. [Google Scholar] [CrossRef]
- Takhviji, V.; Zibara, K.; Azarkeivan, A.; Mehrvar, N.; Mehrvar, N.; Mezginejad, F.; Khosravi, A. Fertility and pregnancy in Iranian thalassemia patients: An update on transfusion complications. Transfus. Med. 2020, 30, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Elalfy, M.S.; Ibrahim, A.S.; Ibrahim, G.S.; Hussein, H.M.A.G.; Mohammed, H.G.E.; Ebeid, F.S.E. Hidden brain iron content in sickle cell disease: Impact on neurocognitive functions. Eur. J. Nucl. Med. Mol. Imaging 2021, 180, 2677–2686. [Google Scholar] [CrossRef]
- Moffitt, T.; Brammer, G.L.; Caspi, A.; Fawcett, J.; Raleigh, M.; Yuwiler, A.; Silva, P. Whole Blood Serotonin Relates to Violence in an Epidemiological Study. Biol. Psychiatry 1998, 43, 446–457. [Google Scholar] [CrossRef]
- Mørkholt, A.S.; Wiborg, O.; Nieland, J.G.K.; Nielsen, S.; Nieland, J.D. Blocking of carnitine palmitoyl transferase 1 potently reduces stress-induced depression in rat highlighting a pivotal role of lipid metabolism. Sci. Rep. 2017, 7, 2158. [Google Scholar] [CrossRef] [Green Version]
- Zapata, I.; Serpell, J.A.; Alvarez, C.E. Genetic mapping of canine fear and aggression. BMC Genom. 2016, 17, 572. [Google Scholar] [CrossRef] [Green Version]
- Coulon, M.; Lévy, F.; Ravel, C.; Nowak, R.; Boissy, A. Mild effects of gestational stress and social reactivity on the onset of mother–young interactions and bonding in sheep. Stress 2014, 17, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.; Hennessy, M.; Sachser, N. Domestication affects the structure, development and stability of biobehavioural profiles. Front. Zool. 2015, 12, S19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiric-Campara, M.; Tupkovic, E.; Mazalovic, E.; Karalic, E.; Biscevic, M.; Djelilovic-Vranic, J.; Alajbegovic, A. Correlation of aggressiveness and anxiety in fighting sports. Med. Arch. 2012, 66, 116–121. [Google Scholar] [CrossRef] [Green Version]
- Stroop, J.R. Studies of interference in serial verbal reactions. J. Exp. Psychol. 1935, 18, 643–662. [Google Scholar] [CrossRef]
- Keinan, G.; Friedland, N.; Kahneman, D.; Roth, D. The effect of stress on the suppression of erroneous competing responses. Anxiety Stress. Coping 1999, 12, 455–476. [Google Scholar] [CrossRef]
- Smith, J.R.; Hayman, G.T.; Wang, S.-J.; Laulederkind, S.J.F.; Hoffman, M.J.; Kaldunski, M.L.; Tutaj, M.; Thota, J.; Nalabolu, H.S.; Ellanki, S.L.R.; et al. The Year of the Rat: The Rat Genome Database at 20: A multi-species knowledgebase and analysis platform. Nucleic Acids Res. 2019, 48, D731–D742. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.; Troesch, S.; Steiner, T.; Brocherie, F.; Girard, O.; Saugy, J.J.; Schmitt, L.; Millet, G.P.; Wehrlin, J.P. Do male athletes with already high initial haemoglobin mass benefit from ‘live high-train low’ altitude training? Exp. Physiol. 2018, 103, 68–76. [Google Scholar] [CrossRef]
- Büdefeld, T.; Tobet, S.A.; Majdic, G. Steroidogenic Factor 1 and the Central Nervous System. J. Neuroendocrinol. 2011, 24, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Domenice, S.; Machado, A.Z.; Ferreira, F.M.; Ferraz-de-Souza, B.; Lerario, A.M.; Lin, L.; Nishi, M.Y.; Gomes, N.L.; da Silva, T.E.; Silva, R.B.; et al. Wide spectrum of NR5A1-related phenotypes in 46,XY and 46,XX individuals. Birth Defects Res. Part C Embryo Today 2016, 108, 309–320. [Google Scholar]
- Fu, Y.-X.; Ji, J.; Shan, F.; Li, J.; Hu, R. Human mesenchymal stem cell treatment of premature ovarian failure: New challenges and opportunities. Stem Cell Res. Ther. 2021, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.D.; Bulun, S. Endometriosis and nuclear receptors. Hum. Reprod. Updat. 2019, 25, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Bösch, F.; Angele, M.K.; Chaudry, I.H. Gender differences in trauma, shock and sepsis. Mil. Med. Res. 2018, 5, 35. [Google Scholar] [CrossRef]
- Kumar, R.; Thompson, E. The structure of the nuclear hormone receptors. Steroids 1999, 64, 310–319. [Google Scholar] [CrossRef]
- Hoffmann, S.; Roeth, R.; Diebold, S.; Gogel, J.; Hassel, D.; Just, S.; Rappold, G.A. Identification and Tissue-Specific Characterization of Novel SHOX-Regulated Genes in Zebrafish Highlights SOX Family Members Among Other Genes. Front. Genet. 2021, 12, 688808. [Google Scholar] [CrossRef] [PubMed]
- German, A.; Mesch, G.; Hochberg, Z. People Are Taller in Countries with Better Environmental Conditions. Front. Endocrinol. 2020, 11, 106. [Google Scholar] [CrossRef] [Green Version]
- Upners, E.N.; Jensen, R.B.; Meyts, E.R.; Dunø, M.; Aksglaede, L.; Juul, A. Short stature homeobox-containing gene duplications in 3.7% of girls with tall stature and normal karyotypes. Acta Paediatr. 2017, 106, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Magann, E.F.; Winchester, M.I.; Carter, D.P.; Martin, J.N.; Bass, J.D.; Morrison, J.C. Factors Adversely Affecting Pregnancy Outcome in the Military. Am. J. Perinatol. 1995, 12, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Rosin, J.M.; Abassah-Oppong, S.; Cobb, J. Comparative transgenic analysis of enhancers from the human SHOX and mouse Shox2 genomic regions. Hum. Mol. Genet. 2013, 22, 3063–3076. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| No. | Rat Gene | NCBI Gene ID | Direct, 5′→3′ | Reverse, 5′→3′ |
|---|---|---|---|---|
| i | ii | iii | iv | v |
| DEGs Identifiedin Hypothalamus of Tame versus Aggressive Rats [This Work] | ||||
| 1 | Apobec1 | 25383 | CGCCGCAACATAAGCTCCCGA | TGCTGTGCCTTCCTCCCCAGTTG |
| 2 | Ascl3 | 246301 | CCTCTGCTGCCCTTTTCCAG | ACTTGACTCGCTGCCTCTCT |
| 3 | Defb17 | 641658 | TGGTAGCTTGGACTTGAGGAAAGAA | TGCAGCAGTGTGTTCCAGGTC |
| Reference Genes | ||||
| 4 | B2m | 24223 | GTGTCTCAGTTCCACCCACC | TTACATGTCTCGGTCCCAGG |
| 5 | Hprt1 | 24465 | TCCCAGCGTCGTGATTAGTGA | CCTTCATGACATCTCGAGCAAG |
| 6 | Ppia | 25518 | TTCCAGGATTCATGTGCCAG | CTTGCCATCCAGCCACTC |
| 7 | Rpl30 | 64640 | CATCTTGGCGTCTGATCTTG | TCAGAGTCTGTTTGTACCCC |
| # | Wild Animals | Domestic Animals | Tissue | NDEG | [Ref] |
|---|---|---|---|---|---|
| 1 | aggressive foxes (Vulpes vulpes) | tame foxes (V. vulpes) | pituitary | 327 | [23] |
| 2 | wolves (Canis familiaris) | dogs (C. lupus) | blood | 450 | [24] |
| 3 | wolves (C. lupus) | dogs (C. familiaris) | frontal cortex | 13 | [25] |
| 4 | Boars (Sus scrofa) | pigs (S. scrofa) | frontal cortex | 30 | [25] |
| 5 | cavy (Cavia aperea) | guinea pigs (C. porcellus) | frontal cortex | 883 | [25] |
| 6 | wild rabbits (Oryctolagus cuniculus) | domestic rabbits (O. cuniculus domesticus) | frontal cortex | 17 | [25] |
| 7 | aggressive rats (R. norvegicus) | tame rats (Rattus norvegicus) | frontal cortex | 20 | [25] |
| 8 | boars (S. scrofa) | pigs (S. scrofa) | frontal cortex | 34 | [26] |
| 9 | boars (S. scrofa) | pigs (S. scrofa) | pituitary | 22 | [27] |
| 10 | wild rabbits (O. cuniculus) | domestic rabbits (O. cuniculus domesticus) | parietal-temporal cortex | 216 | [28] |
| 11 | wild rabbits (O. cuniculus) | domestic rabbits (O. cuniculus domesticus) | amygdala | 118 | [28] |
| 12 | wild rabbits (O. cuniculus) | domestic rabbits (O. cuniculus domesticus) | hypothalamus | 43 | [28] |
| 13 | wild rabbits (O. cuniculus) | domestic rabbits (O. cuniculus domesticus) | hippocampus | 100 | [28] |
| 14 | wild chicken (G. gallus) | domestic chicken (Gallus gallus) | pituitary | 474 | [29] |
| Σ | 7 wild animal species: | 7 domestic animal species: | 7 tissues | 2347 | 7 Refs |
| Group | Tame vs. Aggressive Rats |
|---|---|
| Total sequence reads (NCBI SRA, ID = PRJNA668014) | 219,086,104 |
| Reads mapped to reference rat genome RGSC Rnor_6.0, UCSC Rn6, July 2014 (%) | 184,991,379 (84.44%) |
| Expressed genes identified | 14,039 |
| Candidate DEGs identified (p value < 0.05; Fisher’s Z-test) | 1025 |
| Statistically significant DEGs (PADJ < 0.05, Benjamini correction) | 46 |
| Rat Gene | Differential Expression | ||||
|---|---|---|---|---|---|
| No. | Symbol | Name | log2 | p | PADJ |
| 1 | Ascl3 | achaete-scute family bHLH transcription factor 3 | 3.99 | 10−12 | 10−8 |
| 2 | Morn1 | MORN repeat containing 1 | 1.24 | 10−10 | 10−6 |
| 3 | Krt2 | keratin 2 | −1.65 | 10−8 | 10−4 |
| 4 | Banp | Btg3 associated nuclear protein | −0.82 | 10−6 | 10−3 |
| 5 | Mre11 | MRE11 homolog, double strand break repair nuclease | −2.47 | 10−6 | 10−3 |
| 6 | Rbm3 | RNA binding motif protein 3 | 1.04 | 10−6 | 10−3 |
| 7 | Fcgr3a | Fc fragment of IgG receptor IIIa | 2.06 | 10−6 | 10−2 |
| 8 | Plac8 | placenta associated 8 (synonym: onzin) | 2.83 | 10−5 | 10−2 |
| 9 | Cd22 | CD22 molecule | 2.85 | 10−5 | 10−2 |
| 10 | Apobec1 | apolipoprotein B mRNA editing enzyme catalytic subunit 1 | 1.87 | 10−5 | 10−2 |
| 11 | Magee2 | MAGE family member E2 | −0.95 | 10−5 | 10−2 |
| 12 | Hbb-b1 | hemoglobin, β adult major chain | −3.97 | 10−5 | 10−2 |
| 13 | Tpm2 | tropomyosin 2 | 1.44 | 10−5 | 10−2 |
| 14 | Apobr | apolipoprotein B receptor | 1.56 | 10−5 | 10−2 |
| 15 | Cenps | centromere protein S | 1.63 | 10−4 | 0.05 |
| 16 | Gale | UDP-galactose-4-epimerase | 1.15 | 10−4 | 0.05 |
| 17 | Pcdhb9 | protocadherin β9 | −1.01 | 10−4 | 0.05 |
| 18 | P2rx4 | purinergic receptor P2X 4 | 1.14 | 10−4 | 0.05 |
| 19 | Rn45s | 45S pre-ribosomal RNA | −1.62 | 10−4 | 0.05 |
| 20 | Nr4a3 | nuclear receptor subfamily 4, group A, member 3 | −1.29 | 10−4 | 0.05 |
| 21 | Ghitm | growth hormone inducible transmembrane protein | 0.40 | 10−4 | 0.05 |
| 22 | Shox2 | short stature homeobox 2 | 6.18 | 10−4 | 0.05 |
| 23 | Insig1 | insulin induced gene 1 | 0.49 | 10−4 | 0.05 |
| 24 | Orai1 | ORAI calcium release-activated calcium modulator 1 | 1.83 | 10−4 | 0.05 |
| 25 | Thrsp | thyroid hormone responsive | 1.43 | 10−4 | 0.05 |
| 26 | Spint1 | serine peptidase inhibitor, Kunitz type 1 | −0.91 | 10−4 | 0.05 |
| 27 | Liph | lipase H | 3.28 | 10−4 | 0.05 |
| 28 | Pla2g2c | phospholipase A2, group IIC | −1.08 | 10−4 | 0.05 |
| 29 | Lilrb3l | leukocyte immunoglobulin-like receptor, subfamily B, member 3-like | 7.34 | 10−4 | 0.05 |
| 30 | Hspa1b | heat shock protein family A (Hsp70) member 1B | −1.25 | 10−4 | 0.05 |
| 31 | Nmral1 | NmrA-like redox sensor 1 | 1.18 | 10−4 | 0.05 |
| 32 | Mogat2 | monoacylglycerol O-acyltransferase 2 | 2.08 | 10−4 | 0.05 |
| 33 | Defb17 | defensin β 17 | 6.02 | 10−4 | 0.05 |
| 34 | Sh3bgr | SH3 domain binding glutamate-rich protein | 1.11 | 10−4 | 0.05 |
| 35 | Eif2b3 | eukaryotic translation initiation factor 2B subunit γ | 0.63 | 10−4 | 0.05 |
| 36 | Fcrl2 | Fc receptor-like 2 | 1.12 | 10−4 | 0.05 |
| 37 | Fuca1 | α-L-fucosidase 1 | 1.10 | 10−4 | 0.05 |
| 38 | Bdh1 | 3-hydroxybutyrate dehydrogenase 1 | 0.37 | 10−4 | 0.05 |
| 39 | Rps16 | ribosomal protein S16 | 1.32 | 10−3 | 0.05 |
| 40 | Ifi27l2b | interferon-α-inducible protein 27 like 2B | 2.36 | 10−3 | 0.05 |
| 41 | Ifi47 | interferon-γ-inducible protein 47 | 1.47 | 10−3 | 0.05 |
| 42 | Mcm10 | minichromosome maintenance 10 replication initiation factor | −1.98 | 10−3 | 0.05 |
| 43 | Fjx1 | four-jointed box kinase 1 | 0.83 | 10−3 | 0.05 |
| 44 | Zmym6 | zinc finger MYM-type containing 6 | −0.59 | 10−3 | 0.05 |
| 45 | Use1 | unconventional SNARE in the ER 1 | 1.11 | 10−3 | 0.05 |
| 46 | Fus | FUS RNA-binding protein | 0.48 | 10−3 | 0.05 |
| Study Design | Behavioral “Glove” Test [20] and the qPCR Data on Gene Expression [This Work] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rat | No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Set | |||||||||||
| Glove Test | A | −3 | −3 | −3 | −3 | −3 | −3 | −3 | −3 | ||
| T | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |||
| DEG | Set | Relative Expression with Respect to Four Reference Genes, qPCR, M0 ± SEM | TOTAL | ||||||||
| Ascl3 | A | 0.22 ± 0.04 | 0.14 ± 0.03 | 1.04 ± 0.12 | 0.11 ± 0.03 | 0.22 ± 0.04 | 0.14 ± 0.02 | 0.12 ± 0.03 | 0.12 ± 0.03 | 0.26 ± 0.12 | |
| T | 1.05 ± 0.28 | 1.35 ± 0.35 | 2.05 ± 0.11 | 1.95 ± 0.32 | 2.35 ± 0.24 | 2.61 ± 0.32 | 2.08 ± 0.61 | 2.86 ± 1.10 | 2.04 ± 0.52 | ||
| Apobec1 | A | 1.83 ±0.19 | 0.71 ± 0.27 | 0.30 ± 0.08 | 0.58 ± 0.17 | 0.22 ± 0.09 | 0.58 ± 0.14 | 1.86 ± 0.28 | 0.93 ± 0.16 | 0.88 ± 0.28 | |
| T | 2.09 ± 0.62 | 9.22 ± 0.15 | 1.12 ± 0.05 | 1.12 ± 0.07 | 4.03 ± 0.73 | 3.83 ± 0.08 | 3.49 ± 1.13 | 0.45 ± 0.11 | 3.17 ± 1.07 | ||
| Defb17 | A | 0.01 ± 0.01 | 0.01 ± 0.01 | ND | ND | ND | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | |
| T | 1.57 ± 0.22 | 1.70 ± 0.07 | 2.12 ± 0.51 | 1.20 ± 0.35 | 0.66 ± 0.04 | 1.51 ± 0.56 | 0.90 ± 0.04 | 0.99 ± 0.06 | 1.33 ± 0.35 | ||
| # | Hypothalamic DEGs, Tame vs. Aggressive Rats | DEGs Within the Tissues of the Domestic Animals versus Their Wild Congeners | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DEG | log2 | PADJ | Tame/Domestic | Wild/Aggressive | Tissue | DEG | log2 | PADJ | [Ref] | |
| i | ii | iii | iv | v | vi | vii | viii | ix | x | xi |
| 1 | Banp | −0.82 | 10−3 | rabbits | rabbits | hippocampus | Banp | −1.39 | 0.05 | [28] |
| 2 | Banp | −0.82 | 10−3 | rabbits | rabbits | parietal-temporal cortex | Banp | −1.21 | 10−2 | [28] |
| 3 | Cd22 | 2.85 | 10−2 | dogs | wolves | blood | Cd22 | 2.34 | 0.05 | [24] |
| 4 | Cd22 | 2.85 | 10−2 | foxes | foxes | pituitary | Cd22 | 0.32 | 10−2 | [23] |
| 5 | Defb17 | 6.02 | 0.05 | rabbits | rabbits | parietal-temporal cortex | Defb1 | 1.19 | 10−2 | [28] |
| 6 | Eif2b3 | 0.63 | 0.05 | guinea pigs | cavy | frontal cortex | Eif2b3 | 0.72 | 10−3 | [25] |
| 7 | Fcgr3a | 2.06 | 10−2 | rabbits | rabbits | parietal-temporal cortex | Fcgr3b | 1.35 | 10−2 | [28] |
| 8 | Fcrl2 | 1.12 | 0.05 | foxes | foxes | pituitary | Fcrl1 | 0.43 | 10−2 | [23] |
| 9 | Ghitm | 0.40 | 0.05 | guinea pigs | cavy | frontal cortex | Ghitm | −0.58 | 0.05 | [25] |
| 10 | Hbb-b1 | −3.97 | 10−2 | dogs | wolves | blood | Hbbl | −5.92 | 10−8 | [24] |
| 11 | Hbb-b1 | −3.97 | 10−2 | dogs | wolves | blood | Hba1 | −4.06 | 10−5 | [24] |
| 12 | Hbb-b1 | −3.97 | 10−2 | chicken | chicken | pituitary | Hbad | −1.07 | 10−2 | [29] |
| 13 | Hbb-b1 | −3.97 | 10−2 | dogs | wolves | blood | Hbm | −6.46 | 10−6 | [24] |
| 14 | Hbb-b1 | −3.97 | 10−2 | dogs | wolves | blood | Hbz1 | −7.10 | 10−2 | [24] |
| 15 | Hspa1b | −1.25 | 0.05 | rabbits | rabbits | parietal-temporal cortex | Hspa5 | −1.12 | 0.05 | [28] |
| 16 | Hspa1b | −1.25 | 0.05 | rabbits | rabbits | amygdala | Hspa5 | −1.12 | 0.05 | [28] |
| 17 | Hspa1b | −1.25 | 0.05 | rabbits | rabbits | parietal-temporal cortex | Hspa8 | −1.46 | 10−9 | [28] |
| 18 | Hspa1b | −1.25 | 0.05 | rabbits | rabbits | amygdala | Hspa8 | −1.10 | 0.05 | [28] |
| 19 | Hspa1b | −1.25 | 0.05 | rabbits | rabbits | hippocampus | Hspa8 | −1.36 | 10−2 | [28] |
| 20 | Ifi27l2b | 2.36 | 0.05 | chicken | chicken | pituitary | Ifi6 | −2.49 | 10−4 | [29] |
| 21 | Krt2 | −1.65 | 10−4 | chicken | chicken | pituitary | Krt17 | −1.12 | 0.05 | [29] |
| 22 | Liph | 3.28 | 0.05 | guinea pigs | cavy | frontal cortex | Lipa | 0.84 | 10−2 | [25] |
| 23 | Liph | 3.28 | 0.05 | guinea pigs | cavy | frontal cortex | Lipm | 1.45 | 10−2 | [25] |
| 24 | Liph | 3.28 | 0.05 | chicken | chicken | pituitary | Lipml | 0.55 | 10−3 | [29] |
| 25 | Mogat2 | 2.08 | 0.05 | rabbits | rabbits | hippocampus | Mogat1 | −1.93 | 0.05 | [28] |
| 26 | Morn1 | 1.24 | 10−6 | foxes | foxes | pituitary | Morn2 | −0.25 | 0.05 | [23] |
| 27 | Morn1 | 1.24 | 10−6 | guinea pigs | cavy | frontal cortex | Morn2 | 0.89 | 0.05 | [25] |
| 28 | Mre11a | −2.47 | 10−3 | guinea pigs | cavy | frontal cortex | Mre11a | 1.42 | 10−4 | [25] |
| 29 | Nr4a3 | −1.29 | 10−4 | chicken | chicken | pituitary | Nr4a3 | −0.85 | 0.05 | [29] |
| 30 | Nr4a3 | −1.29 | 10−4 | rabbits | rabbits | amygdala | Nr4a3 | −1.58 | 0.05 | [28] |
| 31 | Nr4a3 | −1.29 | 10−4 | guinea pigs | cavy | frontal cortex | Nr2c1 | −0.74 | 10−2 | [25] |
| 32 | Nr4a3 | −1.29 | 10−4 | chicken | chicken | pituitary | Nr3c1 | 0.51 | 10−5 | [29] |
| 33 | Nr4a3 | −1.29 | 10−4 | guinea pigs | cavy | frontal cortex | Nr5a1 | −2.19 | 0.05 | [25] |
| 34 | Orai1 | 1.83 | 0.05 | guinea pigs | cavy | frontal cortex | Orai1 | −1.30 | 10−3 | [25] |
| 35 | P2rx4 | 1.14 | 0.05 | guinea pigs | cavy | frontal cortex | P2rx6 | 0.55 | 0.05 | [25] |
| 36 | Pcdhb9 | −1.01 | 0.054 | guinea pigs | cavy | frontal cortex | Pcdh20 | −0.73 | 0.05 | [25] |
| 37 | Pcdhb9 | −1.01 | 0.054 | guinea pigs | cavy | frontal cortex | Pcdhac1 | 0.72 | 10−2 | [25] |
| 38 | Pcdhb9 | −1.01 | 0.054 | rabbits | rabbits | parietal-temporal cortex | Pcdhb15 | −1.04 | 0.05 | [28] |
| 39 | Pcdhb9 | −1.01 | 0.054 | rats | rats | frontal cortex | Pcdhga1 | 2.10 | 10−5 | [25] |
| 40 | Pcdhb9 | −1.01 | 0.054 | rabbits | rabbits | amygdala | Pcdhgb4 | 1.53 | 10−4 | [28] |
| 41 | Pcdhb9 | −1.01 | 0.054 | rabbits | rabbits | parietal-temporal cortex | Pcdhgb4 | 1.06 | 10−4 | [28] |
| 42 | Pcdhb9 | −1.01 | 0.054 | rabbits | rabbits | hypothalamus | Pcdhgb4 | 1.67 | 10−2 | [28] |
| 43 | Pla2g2c | −1.08 | 0.05 | rabbits | rabbits | parietal-temporal cortex | Pla1a | 1.35 | 10−2 | [28] |
| 44 | Pla2g2c | −1.08 | 0.05 | guinea pigs | cavy | frontal cortex | Pla2g4a | −1.74 | 10−7 | [25] |
| 45 | Pla2g2c | −1.08 | 0.05 | rabbits | rabbits | parietal-temporal cortex | Pla2g4c | 2.29 | 10−8 | [28] |
| 46 | Pla2g2c | −1.08 | 0.05 | rabbits | rabbits | amygdala | Pla2g4c | 2.34 | 10−3 | [28] |
| 47 | Pla2g2c | −1.08 | 0.05 | rabbits | rabbits | hippocampus | Pla2g4c | 1.63 | 0.05 | [28] |
| 48 | Pla2g2c | −1.08 | 0.05 | guinea pigs | cavy | frontal cortex | Pla2g5 | −1.01 | 0.05 | [25] |
| 49 | Pla2g2c | −1.08 | 0.05 | chicken | chicken | pituitary | Pla2g7 | −0.83 | 10−2 | [29] |
| 50 | Rbm3 | 1.04 | 10−3 | guinea pigs | cavy | frontal cortex | Rbm11 | 1.02 | 0.05 | [25] |
| 51 | Sh3bgr | 1.11 | 0.05 | guinea pigs | cavy | frontal cortex | Sh3bgr | 0.99 | 10−2 | [25] |
| 52 | Shox2 | 6.18 | 0.05 | rabbits | rabbits | hippocampus | Shox2 | −3.43 | 10−3 | [28] |
| 53 | Spint1 | −0.91 | 0.05 | dogs | wolves | blood | Spint1 | 5.28 | 10−2 | [24] |
| 54 | Tpm2 | 1.44 | 10−2 | foxes | foxes | pituitary | Tpm4 | 0.84 | 10−8 | [23] |
| log2 Value | Signs of Log2 Values of Homologous DEGs | Binomial Distribution | χ2 Test | Fisher’s Exact Test | |||
|---|---|---|---|---|---|---|---|
| Homology Type | PC1: Same Signs | PC2: Opposite Signs | χ2 | p | |||
| i | ii | iii | iv | v | vi | vii | viii |
| Number of Orthologous DEG Pairs | Many Homologs | 7 | 0 | 10−3 | 7.78 | 10−2 | 0.05 |
| Only One Gene | 2 | 5 | 0.23 | ||||
| Human | Effect of Gene Expression Changes on Human Reproductive Potential | Binomial Distribution | Pearson’s χ2 -Test | Fisher’s Exact Test | |||
|---|---|---|---|---|---|---|---|
| Animals | Decreased (→) | Increased (←) | χ2 | p | |||
| i | ii | iii | iv | v | vii | viii | ix |
| Gene Expression Changes During Divergence from Most Recent Common Ancestor | domestic | 13 | 1 | 10−3 | 6.30 | 0.05 | 0.05 |
| wild | 7 | 7 | 0.60 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chadaeva, I.; Ponomarenko, P.; Kozhemyakina, R.; Suslov, V.; Bogomolov, A.; Klimova, N.; Shikhevich, S.; Savinkova, L.; Oshchepkov, D.; Kolchanov, N.A.; et al. Domestication Explains Two-Thirds of Differential-Gene-Expression Variance between Domestic and Wild Animals; The Remaining One-Third Reflects Intraspecific and Interspecific Variation. Animals 2021, 11, 2667. https://doi.org/10.3390/ani11092667
Chadaeva I, Ponomarenko P, Kozhemyakina R, Suslov V, Bogomolov A, Klimova N, Shikhevich S, Savinkova L, Oshchepkov D, Kolchanov NA, et al. Domestication Explains Two-Thirds of Differential-Gene-Expression Variance between Domestic and Wild Animals; The Remaining One-Third Reflects Intraspecific and Interspecific Variation. Animals. 2021; 11(9):2667. https://doi.org/10.3390/ani11092667
Chicago/Turabian StyleChadaeva, Irina, Petr Ponomarenko, Rimma Kozhemyakina, Valentin Suslov, Anton Bogomolov, Natalya Klimova, Svetlana Shikhevich, Ludmila Savinkova, Dmitry Oshchepkov, Nikolay A. Kolchanov, and et al. 2021. "Domestication Explains Two-Thirds of Differential-Gene-Expression Variance between Domestic and Wild Animals; The Remaining One-Third Reflects Intraspecific and Interspecific Variation" Animals 11, no. 9: 2667. https://doi.org/10.3390/ani11092667
APA StyleChadaeva, I., Ponomarenko, P., Kozhemyakina, R., Suslov, V., Bogomolov, A., Klimova, N., Shikhevich, S., Savinkova, L., Oshchepkov, D., Kolchanov, N. A., Markel, A., & Ponomarenko, M. (2021). Domestication Explains Two-Thirds of Differential-Gene-Expression Variance between Domestic and Wild Animals; The Remaining One-Third Reflects Intraspecific and Interspecific Variation. Animals, 11(9), 2667. https://doi.org/10.3390/ani11092667






