Developmental Change of Yolk Microbiota and Its Role on Early Colonization of Intestinal Microbiota in Chicken Embryo
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Animals and Sample Preparation
2.2. DNA Extraction and 16s rRNA Sequencing
2.3. Sequence Analysis
2.4. Bioinformatics and Statistical Analysis
3. Results
3.1. Composition of Microbiota in Yolk and Gut of Chicken Embryo
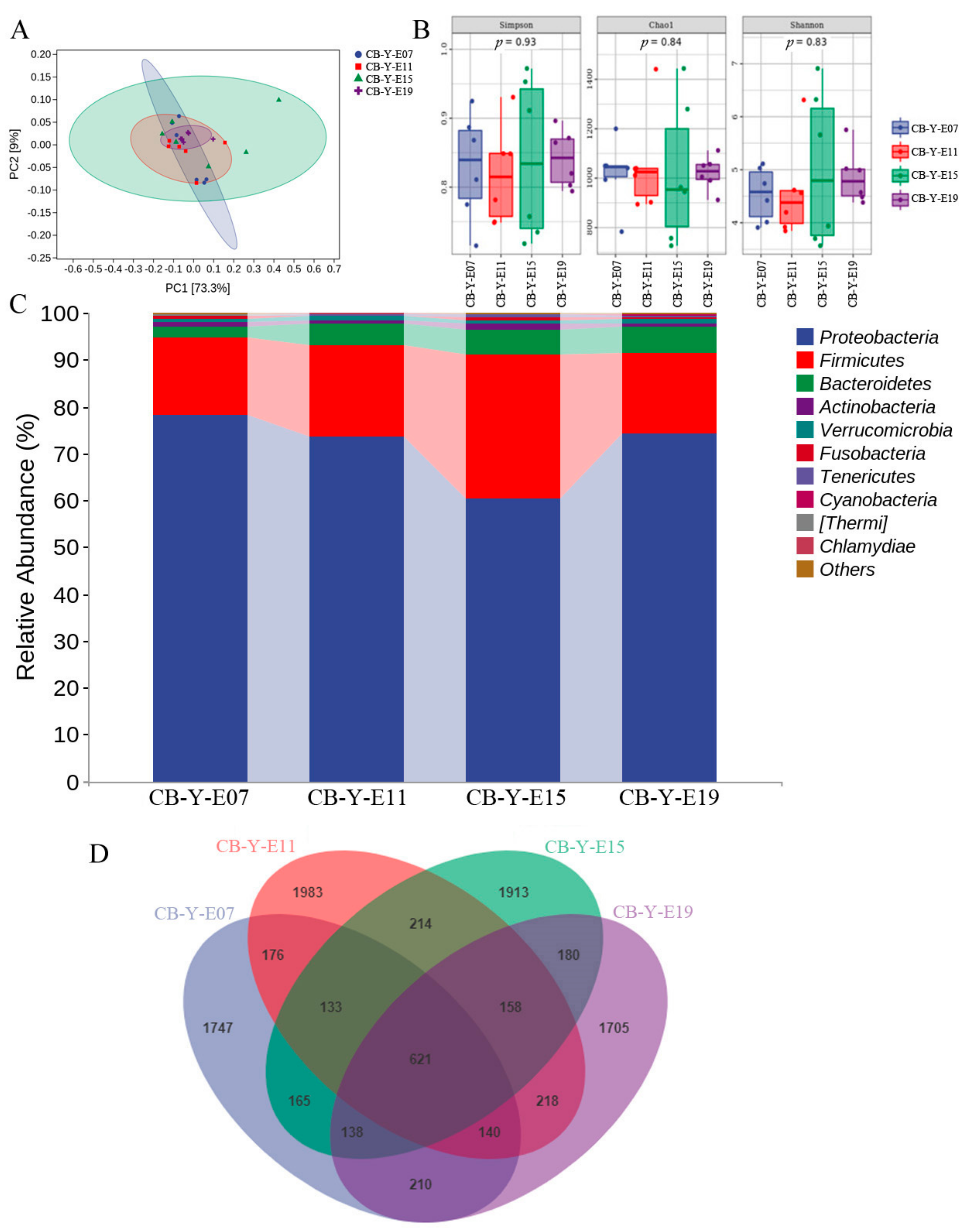
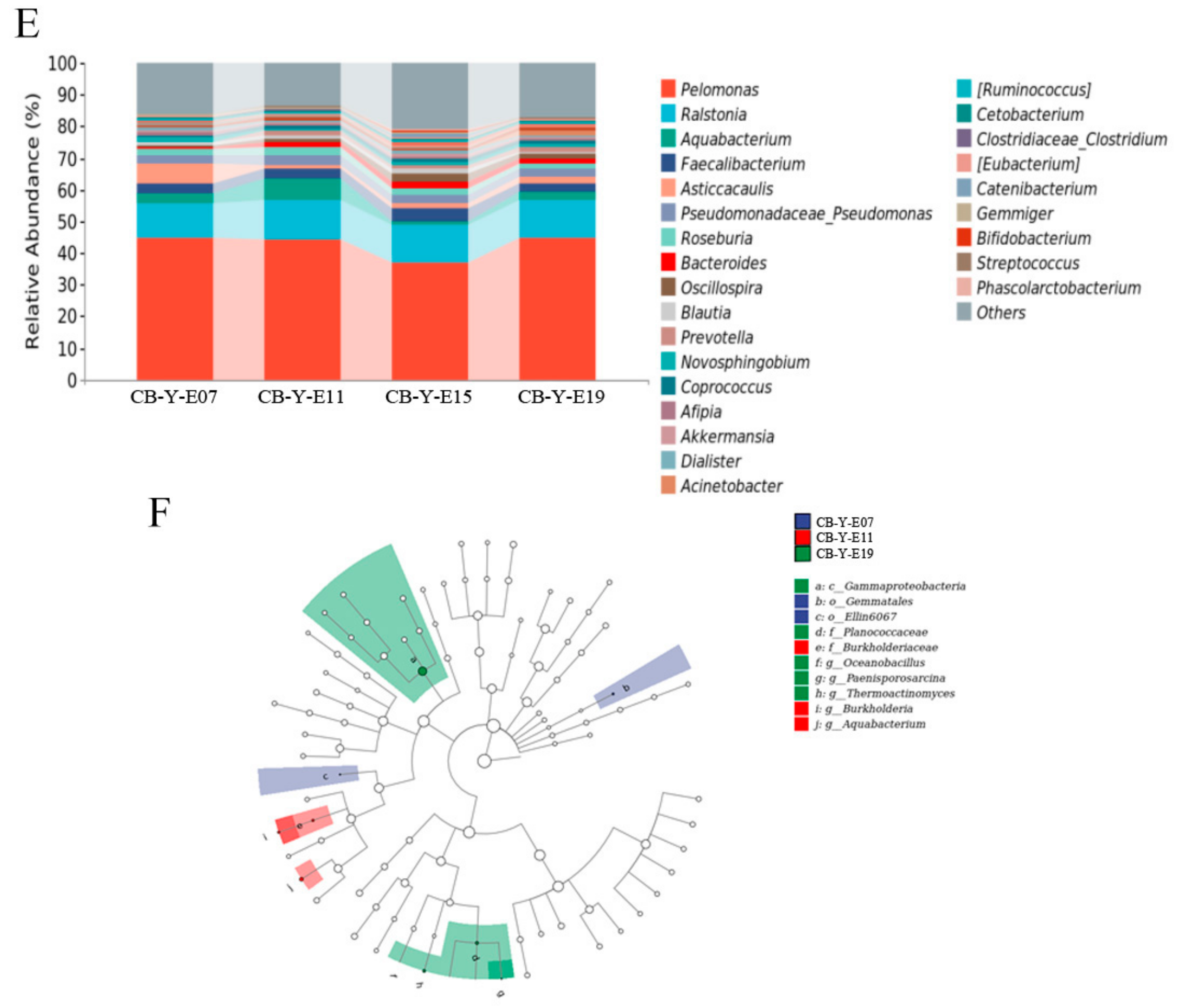
3.2. Change of Yolk Microbiota at Different Developmental Stages
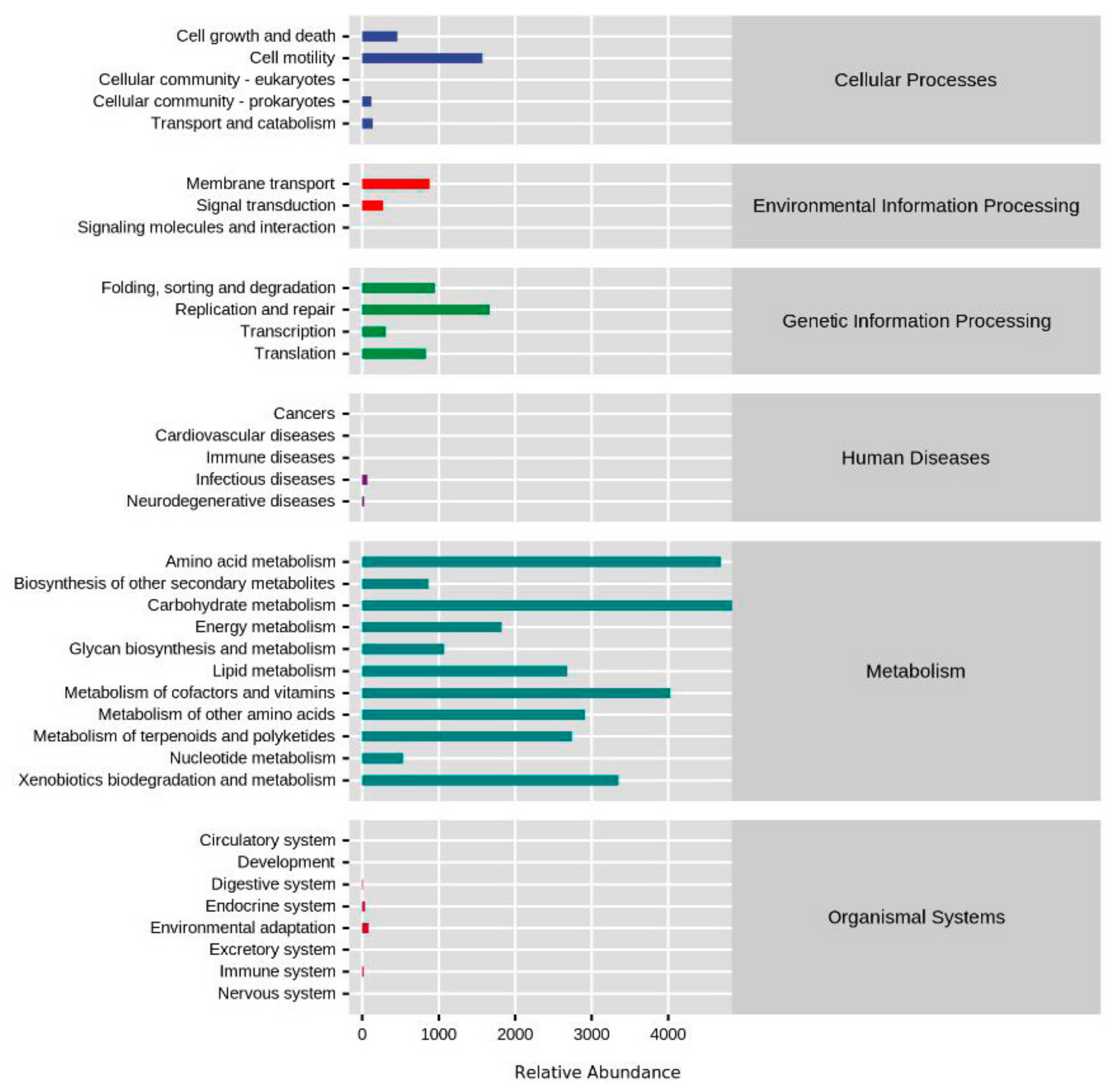
3.3. Functional Profiles of Yolk Microbial Community during Embryogenesis
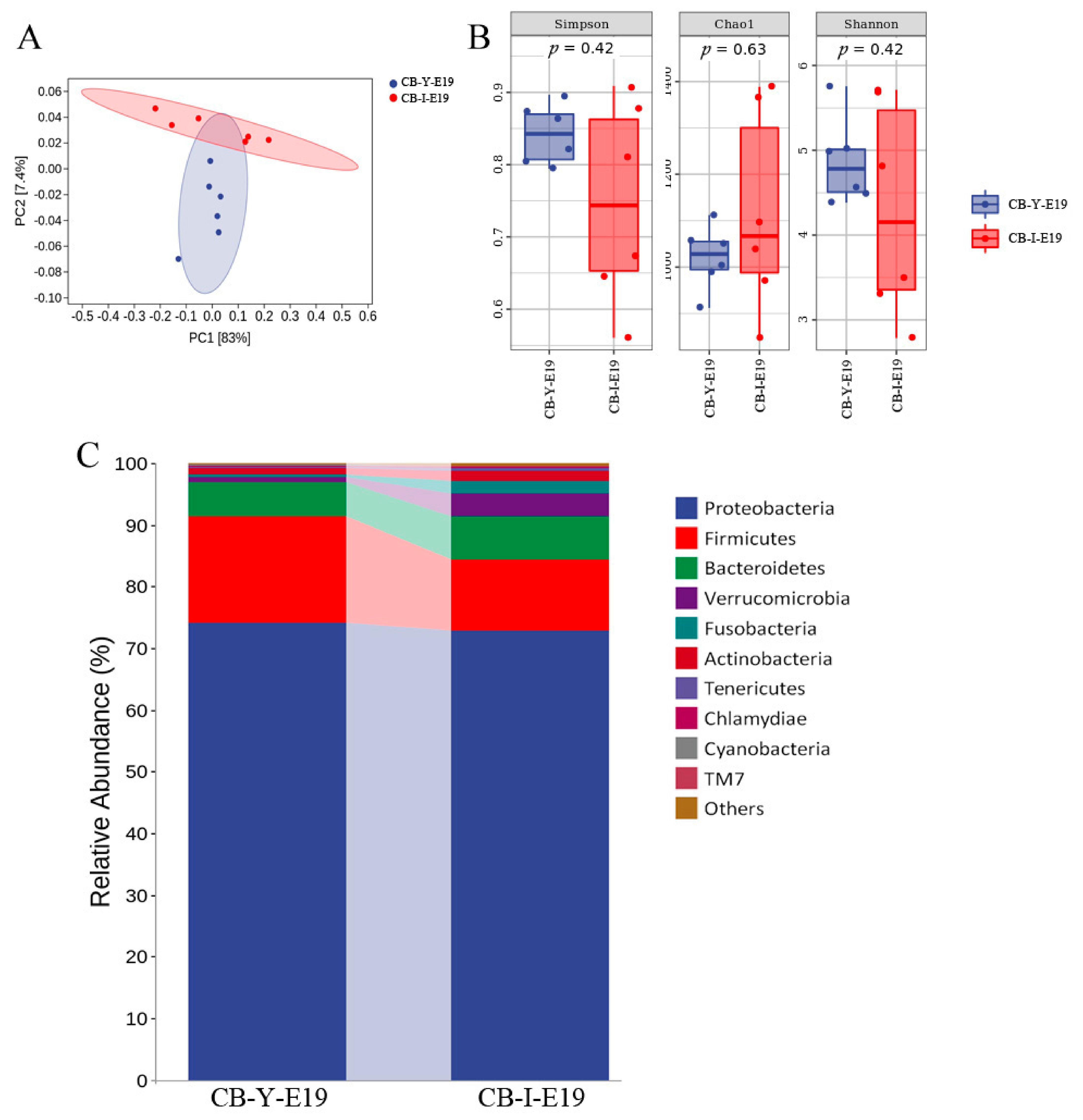
3.4. Comparison of the Microbiota between Yolk and Intestine at E19
3.5. Transition of Intestinal Microbiota from Late Embryonic to Early Posthatch Stage
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Doyle, R.M.; Alber, D.G.; Jones, H.E.; Harris, K.; Fitzgerald, F.; Peebles, D.; Klein, N. Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta 2014, 35, 1099–1101. [Google Scholar] [CrossRef] [Green Version]
- Walther-Antonio, M.R.; Chen, J.; Multinu, F.; Hokenstad, A.; Distad, T.J.; Cheek, E.H.; Keeney, G.L.; Creedon, D.J.; Nelson, H.; Mariani, A.; et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, I.; Corwin, E.J.; Brennan, P.A.; Jordan, S.; Murphy, J.R.; Dunlop, A. The Infant Microbiome: Implications for Infant Health and Neurocognitive Development. Nurs. Res. 2016, 65, 76–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, S.L. Chicken embryo utilization of egg micronutrients. Braz. J. Poult. Sci. 2007, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bauer, R.; Plieschnig, J.A.; Finkes, T.; Riegler, B.; Hermann, M.; Schneider, W.J. The developing chicken yolk sac acquires nutrient transport competence by an orchestrated differentiation process of its endodermal epithelial cells. J. Biol. Chem. 2013, 288, 1088–1098. [Google Scholar] [CrossRef] [Green Version]
- Yadgary, L.; Wong, E.A.; Uni, Z. Temporal transcriptome analysis of the chicken embryo yolk sac. BMC Genom. 2014, 15, 690. [Google Scholar] [CrossRef] [Green Version]
- Speier, J.S.; Yadgary, L.; Uni, Z.; Wong, E.A. Gene expression of nutrient transporters and digestive enzymes in the yolk sac membrane and small intestine of the developing embryonic chick. Poult. Sci. 2012, 91, 1941–1949. [Google Scholar] [CrossRef]
- Zhang, H.; Wong, E.A. Spatial transcriptional profile of PepT1 mRNA in the yolk sac and small intestine in broiler chickens. Poult. Sci. 2017, 96, 2871–2876. [Google Scholar] [CrossRef]
- Van der Wagt, I.; de Jong, I.C.; Mitchell, M.A.; Molenaar, R.; van den Brand, H. A review on yolk sac utilization in poultry. Poult. Sci. 2020, 99, 2162–2175. [Google Scholar] [CrossRef]
- Akinyemi, F.T.; Ding, J.; Zhou, H.; Xu, K.; He, C.; Han, C.; Zheng, Y.; Luo, H.; Yang, K.; Gu, C.; et al. Dynamic distribution of gut microbiota during embryonic development in chicken. Poult. Sci. 2020, 99, 5079–5090. [Google Scholar] [CrossRef]
- Cisek, A.A.; Binek, M. Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol. J. Vet. Sci. 2014, 17, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Cho, S.; La, T.M.; Lee, H.J.; Lee, J.B.; Park, S.Y.; Song, C.S.; Choi, I.S.; Lee, S.W. Comparison of microbiota in the cloaca, colon, and magnum of layer chicken. PLoS ONE 2020, 15, e237108. [Google Scholar] [CrossRef]
- Lee, S.; La, T.M.; Lee, H.J.; Choi, I.S.; Song, C.S.; Park, S.Y.; Lee, J.B.; Lee, S.W. Characterization of microbial communities in the chicken oviduct and the origin of chicken embryo gut microbiota. Sci. Rep. 2019, 9, 6838. [Google Scholar] [CrossRef]
- Cuperus, T.; Kraaij, M.D.; Zomer, A.L.; van Dijk, A.; Haagsman, H.P.; Han, X. Immunomodulation and effects on microbiota after in ovo administration of chicken cathelicidin-2. PLoS ONE 2018, 13, e198188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunislawska, A.; Slawinska, A.; Bednarczyk, M.; Siwek, M. Transcriptome modulation by in ovo delivered Lactobacillus synbiotics in a range of chicken tissues. Gene 2019, 698, 27–33. [Google Scholar] [CrossRef]
- Siwek, M.; Slawinska, A.; Stadnicka, K.; Bogucka, J.; Dunislawska, A.; Bednarczyk, M. Prebiotics and synbiotics—In ovo delivery for improved lifespan condition in chicken. BMC Vet. Res. 2018, 14, 402. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 2014, 5, 508. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Desantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brian, H.M.; Marti, J.A. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asnicar, F.; Weingart, G.; Tickle, T.L.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015, 3, e1029. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [Green Version]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega, T.R.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.; Dai, R.; Yang, L.; He, C.; Xu, K.; Liu, S.; Zhao, W.; Xiao, L.; Luo, L.; Zhang, Y.; et al. Inheritance and Establishment of Gut Microbiota in Chickens. Front. Microbiol. 2017, 8, 1967. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ding, P.; Tong, Y.; He, X.; Yin, Y.; Zhang, H.; Song, Z. Metabolomic analysis of the egg yolk during the embryonic development of broilers. Poult. Sci. 2021, 100, 101014. [Google Scholar] [CrossRef]
- Damms-Machado, A.; Mitra, S.; Schollenberger, A.E.; Kramer, K.M.; Meile, T.; Konigsrainer, A.; Huson, D.H.; Bischoff, S.C. Effects of surgical and dietary weight loss therapy for obesity on gut microbiota composition and nutrient absorption. BioMed Res. Int. 2015, 2015, 806248. [Google Scholar] [CrossRef]
- Gao, P.; Liu, Y.; Le, B.; Qin, B.; Liu, M.; Zhao, Y.; Guo, X.; Cao, G.; Liu, J.; Li, B.; et al. A comparison of dynamic distributions of intestinal microbiota between Large White and Chinese Shanxi Black pigs. Arch. Microbiol. 2019, 201, 357–367. [Google Scholar] [CrossRef]
- Indiani, C.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.; Darrieux, M.; Parisotto, T.M. Childhood Obesity and Firmicutes/Bacteroidetes Ratio in the Gut Microbiota: A Systematic Review. Child Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Yadgary, L.; Kedar, O.; Adepeju, O.; Uni, Z. Changes in yolk sac membrane absorptive area and fat digestion during chick embryonic development. Poult. Sci. 2013, 92, 1634–1640. [Google Scholar] [CrossRef]
- de Faria Ghetti, F.; Oliveira, D.G.; de Oliveira, J.M.; de Castro Ferreira, L.E.V.V.; Cesar, D.E.; Moreira, A.P.B. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur. J. Nutr. 2018, 57, 861–876. [Google Scholar] [CrossRef] [PubMed]
- Zhang-Sun, W.; Terce, F.; Burcelin, R.; Novikov, A.; Serino, M.; Caroff, M. Structure function relationships in three lipids A from the Ralstonia genus rising in obese patients. Biochimie 2019, 159, 72–80. [Google Scholar] [CrossRef] [Green Version]
- Cogburn, L.A.; Trakooljul, N.; Chen, C.; Huang, H.; Wu, C.H.; Carre, W.; Wang, X.; White, H.R. Transcriptional profiling of liver during the critical embryo-to-hatchling transition period in the chicken (Gallus gallus). BMC Genom. 2018, 19, 695. [Google Scholar] [CrossRef]
- Xu, E.; Zhang, L.; Yang, H.; Shen, L.; Feng, Y.; Ren, M.; Xiao, Y. Transcriptome profiling of the liver among the prenatal and postnatal stages in chickens. Poult. Sci. 2019, 98, 7030–7040. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Kidrick, J.; Wong, E.A. Localization of cells expressing SGLT1 mRNA in the yolk sac and small intestine of broilers. Poult. Sci. 2019, 98, 984–990. [Google Scholar] [CrossRef]
- Uni, Z.; Geyra, A.; Ben-Hur, H.; Sklan, D. Small intestinal development in the young chick: Crypt formation and enterocyte proliferation and migration. Brit. Poult. Sci. 2000, 41, 544–551. [Google Scholar] [CrossRef]
- Uni, Z.; Tako, E.; Gal-Garber, O.; Sklan, D. Morphological, molecular, and functional changes in the chicken small intestine of the late-term embryo. Poult. Sci. 2003, 82, 1747–1754. [Google Scholar] [CrossRef]
- Alessi, A.M.; Gray, V.; Farquharson, F.M.; Flores-Lopez, A.; Shaw, S.; Stead, D.; Wegmann, U.; Shearman, C.; Gasson, M.; Collie-Duguid, E.; et al. beta-Glucan is a major growth substrate for human gut bacteria related to Coprococcus eutactus. Environ. Microbiol. 2020, 22, 2150–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Park, J.E.; Lee, K.C.; Choi, S.H.; Oh, B.S.; Yu, S.Y.; Eom, M.K.; Kang, S.W.; Han, K.I.; Suh, M.K.; et al. Blautia faecicola sp. nov., isolated from faeces from a healthy human. Int. J. Syst. Evol. Microbiol. 2020, 70, 2059–2065. [Google Scholar] [CrossRef]
- Del, C.F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandona, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar]
- Ferreira-Halder, C.V.; Faria, A.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, P.; Liu, H.; Tong, Y.; He, X.; Yin, X.; Yin, Y.; Zhang, H.; Song, Z. Developmental Change of Yolk Microbiota and Its Role on Early Colonization of Intestinal Microbiota in Chicken Embryo. Animals 2022, 12, 16. https://doi.org/10.3390/ani12010016
Ding P, Liu H, Tong Y, He X, Yin X, Yin Y, Zhang H, Song Z. Developmental Change of Yolk Microbiota and Its Role on Early Colonization of Intestinal Microbiota in Chicken Embryo. Animals. 2022; 12(1):16. https://doi.org/10.3390/ani12010016
Chicago/Turabian StyleDing, Peng, Huichao Liu, Yueyue Tong, Xi He, Xin Yin, Yulong Yin, Haihan Zhang, and Zehe Song. 2022. "Developmental Change of Yolk Microbiota and Its Role on Early Colonization of Intestinal Microbiota in Chicken Embryo" Animals 12, no. 1: 16. https://doi.org/10.3390/ani12010016
APA StyleDing, P., Liu, H., Tong, Y., He, X., Yin, X., Yin, Y., Zhang, H., & Song, Z. (2022). Developmental Change of Yolk Microbiota and Its Role on Early Colonization of Intestinal Microbiota in Chicken Embryo. Animals, 12(1), 16. https://doi.org/10.3390/ani12010016





