Functional Characterization of Melanocortin-3 Receptor in a Hibernating Cavefish Onychostoma macrolepis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Primers
2.2. Total RNA Extraction and Reverse Transcription (RT)
2.3. Molecular Cloning of omMc3r and Construction of Its Eukaryotic Expression Plasmid
2.4. Homology and Phylogenetic Analyses of omMc3r
2.5. Tissue Expression of omMc3r
2.6. Cell Culture
2.7. Luciferase Reporter Assay
2.8. Statistical Analysis
3. Results
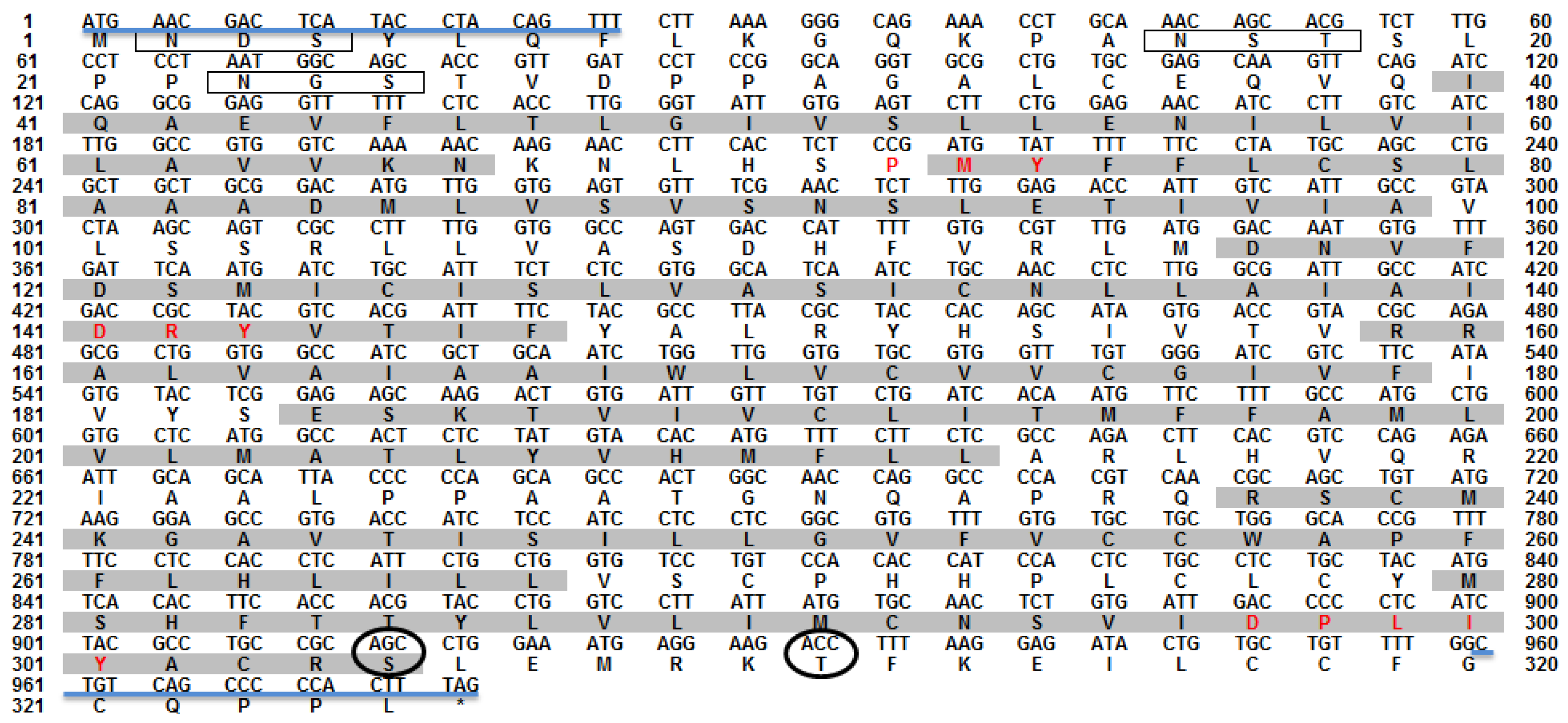
3.1. Nucleotide and Deduced Amio Acid Sequences of omMc3r
3.2. Phylogenetic Analysis
3.3. Tissue Expression of omMc3r
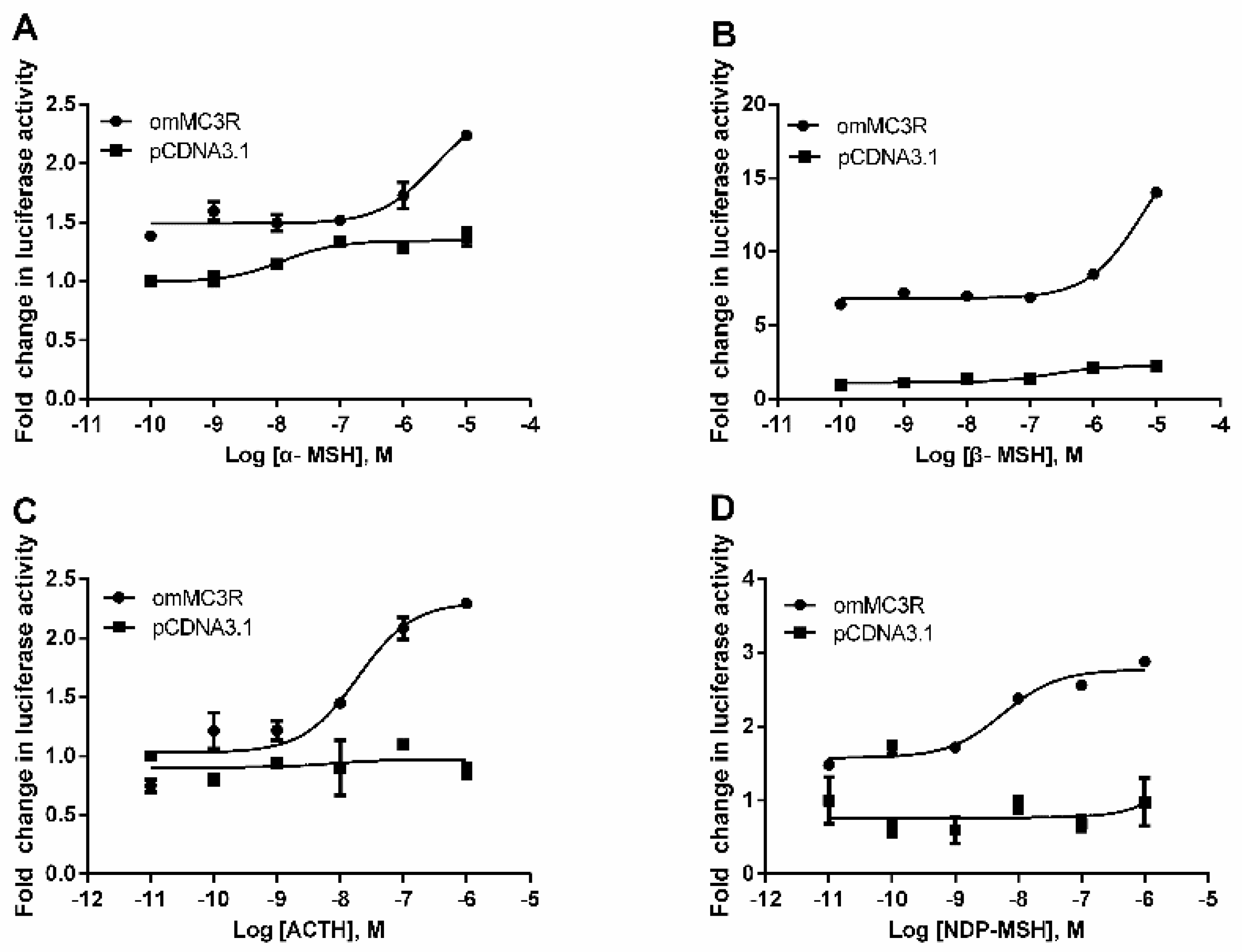
3.4. Functional Characteristics of omMc3r in HEK293T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gantz, I.; Fong, T.M. The melanocortin system. Am. J. Physiol. Metab. 2003, 284, E468–E474. [Google Scholar] [CrossRef]
- Rana, B.K.; Hewett-Emmett, D.; Jin, L.; Chang, B.H.; Sambuughin, N.; Lin, M.; Watkins, S.; Bamshad, M.; Jorde, L.B.; Ramsay, M.; et al. High polymorphism at the human melanocortin 1 receptor locus. Genetics 1999, 151, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Klovins, J.; Haitina, T.; Ringholm, A.; Löwgren, M.; Fridmanis, D.; Slaidina, M.; Stier, S.; Schiöth, H.B. Cloning of two melanocortin (MC) receptors in spiny dogfish: MC3 receptor in cartilaginous fish shows high affinity to ACTH-derived peptides while it has lower preference to γ-MSH. Eur. J. Biochem. 2004, 271, 4320–4331. [Google Scholar] [CrossRef]
- Chhajlani, V. Distribution of cDNA for melanocortin receptor subtypes in human tissues. Biochem. Mol. Biol. Int. 1996, 38, 73. [Google Scholar]
- Chen, W.; Kelly, M.; Opitz-Araya, X.; Thomas, R.; Low, M.; Cone, R.; Chen, W.B.; Kelly, M.A.; Opitz-Araya, X.; Thomas, R.E.; et al. Exocrine gland dysfunction in MC5-R-deficient mice: Evidence for coordinated regulation of exocrine gland function by melanocortin peptides. Cell 1998, 91, 789–798. [Google Scholar] [CrossRef] [Green Version]
- Ericson, M.D.; Doering, S.R.; Larson, C.M.; Freeman, K.T.; LaVoi, T.M.; Donow, H.M.; Santos, R.G.; Cho, R.H.; Koerperich, Z.M.; Giulianotti, M.A.; et al. Functional mixture-based positional scan identifies a library of antagonist tetrapeptide sequences (LAtTeS) with nanomolar potency for the melanocortin-4 receptor and equipotent with the endogenous AGRP(86-132) antagonist. J. Med. Chem. 2021, 64, 14860–14875. [Google Scholar] [CrossRef] [PubMed]
- Dunigan, A.I.; Olson, D.P.; Roseberry, A.G. Neuropharmacology VTA MC3R neurons control feeding in an activity- and sex-dependent manner in mice. Neuropharmacology 2021, 197, 108746. [Google Scholar] [CrossRef] [PubMed]
- Metherell, L.A.; Chapple, J.P.; Cooray, S.; David, A.; Becker, C.; Rüschendorf, F.; Naville, D.; Begeot, M.; Khoo, B.; Nürnberg, P.; et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat. Genet. 2005, 37, 166–170. [Google Scholar] [CrossRef]
- Chan, L.F.; Webb, T.R.; Chung, T.T.; Meimaridou, E.; Cooray, S.N.; Guasti, L.; Chapple, J.P.; Egertová, M.; Elphick, M.R.; Cheetham, M.E.; et al. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. Proc. Natl. Acad. Sci. USA 2009, 106, 6146–6151. [Google Scholar] [CrossRef] [Green Version]
- Girardet, C.; Butler, A.A. Neural melanocortin receptors in obesity and related metabolic disorders. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 482–494. [Google Scholar] [CrossRef] [Green Version]
- Patel, H.; Montero-Melendez, T.; Greco, K.; Perretti, M. Melanocortin receptors as novel effectors of macrophage responses in inflammation. Front. Immunol. 2011, 2, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.K.; Zhang, Z.R.; Wen, H.S.; Tao, Y.X. Characterization of channel catfish (Ictalurus punctatus) melanocortin-3 receptor reveals a potential network in regulation of energy homeostasis. Gen. Comp. Endocrinol. 2019, 277, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tao, Y. Biased signaling at neural melanocortin receptors in regulation of energy homeostasis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2486–2495. [Google Scholar] [CrossRef]
- Chai, B.; Li, J.-Y.; Zhang, W.; Ammori, J.B.; Mulholland, M.W. Melanocortin-3 receptor activates MAP kinase via PI3 kinase. Regul. Pept. 2007, 139, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Klovins, J.; Haitina, T.; Fridmanis, D.; Kilianova, Z.; Kapa, I.; Fredriksson, R.; Gallo-Payet, N.; Schioöth, H.B. The melanocortin system in Fugu: Determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol. Biol. Evol. 2004, 21, 563–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, D.W.; Bryson-Richardson, R.J.; Pagán, K.E.; Taylor, M.S.; Currie, P.D.; Jackson, I.J. The structure and evolution of the melanocortin and MCH receptors in fish and mammals. Genomics 2003, 81, 184–191. [Google Scholar] [CrossRef]
- Selz, Y.; Braasch, I.; Hoffmann, C.; Schmidt, C.; Schultheis, C.; Schartl, M.; Volff, J.N. Evolution of melanocortin receptors in teleost fish: The melanocortin type 1 receptor. Gene 2007, 401, 114–122. [Google Scholar] [CrossRef]
- Liao, S.; Chen, K.; Xi, B.; Qin, T.; Pan, L.; Xie, J. Molecular cloning, characterization, and expression analysis of Megalobrama amblycephala melanocortin receptor 3 during fasting. J. Fish. Sci. China 2019, 26, 445–456. [Google Scholar] [CrossRef]
- Renquist, B.J.; Zhang, C.; Williams, S.Y.; Cone, R.D. Development of an assay for high-throughput energy expenditure monitoring in the zebrafish. Zebrafish 2013, 10, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, A.; Davis, P.; Reinick, C.; Mizusawa, K.; Sakamoto, T.; Dores, R.M. Characterization of melanocortin receptors from stingray Dasyatis akajei, a cartilaginous fish. Gen. Comp. Endocrinol. 2016, 232, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Ji, R.-L.; Huang, L.; Wang, Y.; Liu, T.; Fan, S.-Y.; Tao, M.; Tao, Y.-X. Topmouth culter melanocortin-3 receptor: Regulation by two isoforms of melanocortin-2 receptor accessory protein 2. Endocr. Connect. 2021, 1, 1489–1501. [Google Scholar] [CrossRef]
- Ma, L.; Zhao, Y.; Yang, J. Cavefish of China. In Encyclopedia of Caves, 3rd ed.; White, B.W., Culver, C.D., Eds.; Academic Press: Cambridge, CA, USA, 2019; pp. 237–254. [Google Scholar]
- Aspirasa, A.C.; Rohnera, N.; Martineaua, B.; Borowskyb, R.L.; Tabina, C.J. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl. Acad. Sci. USA 2015, 112, 9668–9673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Huang, H.; Tao, Y.X. Functions of the DRY motif and intracellular loop 2 of human melanocortin 3 receptor. J. Mol. Endocrinol. 2014, 53, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Huang, Z.L.; Tao, Y.X. Functions of DPLIY motif and helix 8 of human melanocortin-3 receptor. J. Mol. Endocrinol. 2015, 55, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Harmon, C.M. Molecular signatures of human melanocortin receptors for ligand binding and signaling. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2017, 1863, 2436–2447. [Google Scholar] [CrossRef]
- Levi, N.J.; Jun, J.; Patel, T.P.; Uhlman, A.J.; Roberson, R.; Yanovski, J.A. Liver-specific reactivation of MC3R partially reverses the obesity of MC3R deficiency. Diabetes 2018, 67, 2017. [Google Scholar] [CrossRef]
- Lee, E.J.; Lee, S.; Jung, J.; Lee, W.; Kim, B.J.; Park, K.W.; Lim, S.; Yoon, C.; Baik, J. Differential regulation of cAMP-mediated gene transcription and ligand selectivity by MC3R and MC4R melanocortin receptors. Eur. J. Biochem. 2001, 268, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.C.; Sartin, J.L.; Tao, Y.X. Molecular cloning and pharmacological characterization of porcine melanocortin-3 receptor. J. Endocrinol. 2008, 196, 139–148. [Google Scholar] [CrossRef]
- Lisak, R.P.; Benjamins, J.A. Melanocortins, melanocortin receptors and multiple sclerosis. Brain Sci. 2017, 7, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.T.; Yang, Z.; Chen, H.P.; Zhu, C.H.; Deng, S.P.; Li, G.L.; Tao, Y.X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in spotted scat, Scatophagus argus. Gen. Comp. Endocrinol. 2016, 230–231, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Dores, R.M.; Londraville, R.L.; Prokop, J.; Davis, P.; Dewey, N.; Lesinski, N. Molecular evolution of GPCRs: Melanocortin/melanocortin receptors. J. Mol. Endocrinol. 2014, 52, 29–42. [Google Scholar] [CrossRef] [Green Version]
- Dores, R.M.; Baron, A.J. Evolution of POMC: Origin, phylogeny, posttranslational processing, and the melanocortins. Ann. N. Y. Acad. Sci. 2011, 1220, 34–48. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequence | Product Size | Ta 1 |
|---|---|---|---|
| omMc3r-F | ATGAACGACTCATACCTACAGTTT | 978 bp | 58 °C |
| omMc3r-R | CTAAAGTGGGGGCTGACAG | ||
| qPCR-F | CTTCTCGCCAGACTTCACG | 100 bp | 55 °C |
| qPCR-R | TCACGGCTCCCTTCATACA | ||
| β-actin-F | TCCACCCGCGAGTACAACCTT | 173 bp | 57 °C |
| β-actin-R | CCCACGATGGAGGGGAAGAC | ||
| gapdh-F | TGTGGGCAAAGTCATTCCTG | 139 bp | 54 °C |
| gapdh-R | GACAGACTCCTTGATGTTAGCGTA |
| Ligand | cAMP Response | ERK1/2 Response |
|---|---|---|
| EC50 (μM) | EC50 (μM) | |
| α-MSH | 0.57 ± 0.00478 | 3.06 ± 0.98 |
| β-MSH | 0.27 ± 0.00495 | 4.85 ± 0.0107 |
| ACTH (1–24) | 0.029 ± 0.00706 | 0.019 ± 0.00128 |
| NDP-MSH | 0.15 ± 0.00197 | 0.0057 ± 0.0012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Yu, H.; Mo, H.; Lan, X.; Pan, C.; Wang, L.; Zhao, H.; Zhou, J.; Li, Y. Functional Characterization of Melanocortin-3 Receptor in a Hibernating Cavefish Onychostoma macrolepis. Animals 2022, 12, 38. https://doi.org/10.3390/ani12010038
Wu L, Yu H, Mo H, Lan X, Pan C, Wang L, Zhao H, Zhou J, Li Y. Functional Characterization of Melanocortin-3 Receptor in a Hibernating Cavefish Onychostoma macrolepis. Animals. 2022; 12(1):38. https://doi.org/10.3390/ani12010038
Chicago/Turabian StyleWu, Lian, Huixia Yu, Haolin Mo, Xianyong Lan, Chuanying Pan, Lixin Wang, Haiyu Zhao, Jishu Zhou, and Yang Li. 2022. "Functional Characterization of Melanocortin-3 Receptor in a Hibernating Cavefish Onychostoma macrolepis" Animals 12, no. 1: 38. https://doi.org/10.3390/ani12010038
APA StyleWu, L., Yu, H., Mo, H., Lan, X., Pan, C., Wang, L., Zhao, H., Zhou, J., & Li, Y. (2022). Functional Characterization of Melanocortin-3 Receptor in a Hibernating Cavefish Onychostoma macrolepis. Animals, 12(1), 38. https://doi.org/10.3390/ani12010038







