Within- and between-Breed Selection Signatures in the Original and Improved Valachian Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Inbreeding Levels
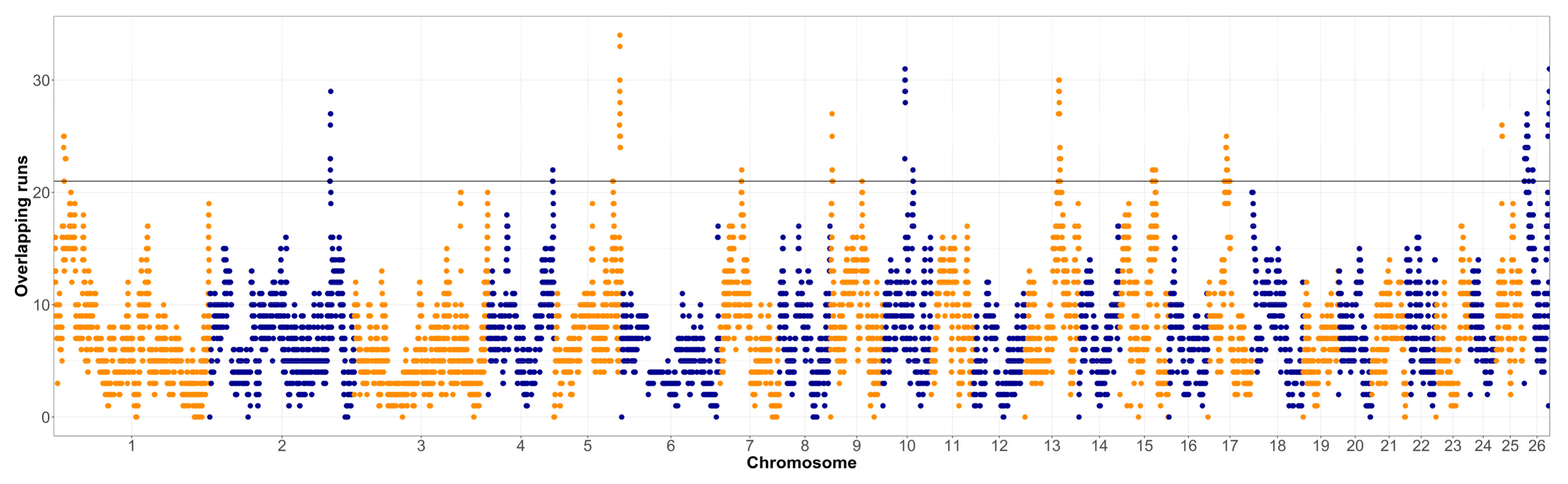
3.2. Within-Breed Selection Signatures Based on Run of Homozygosity Islands
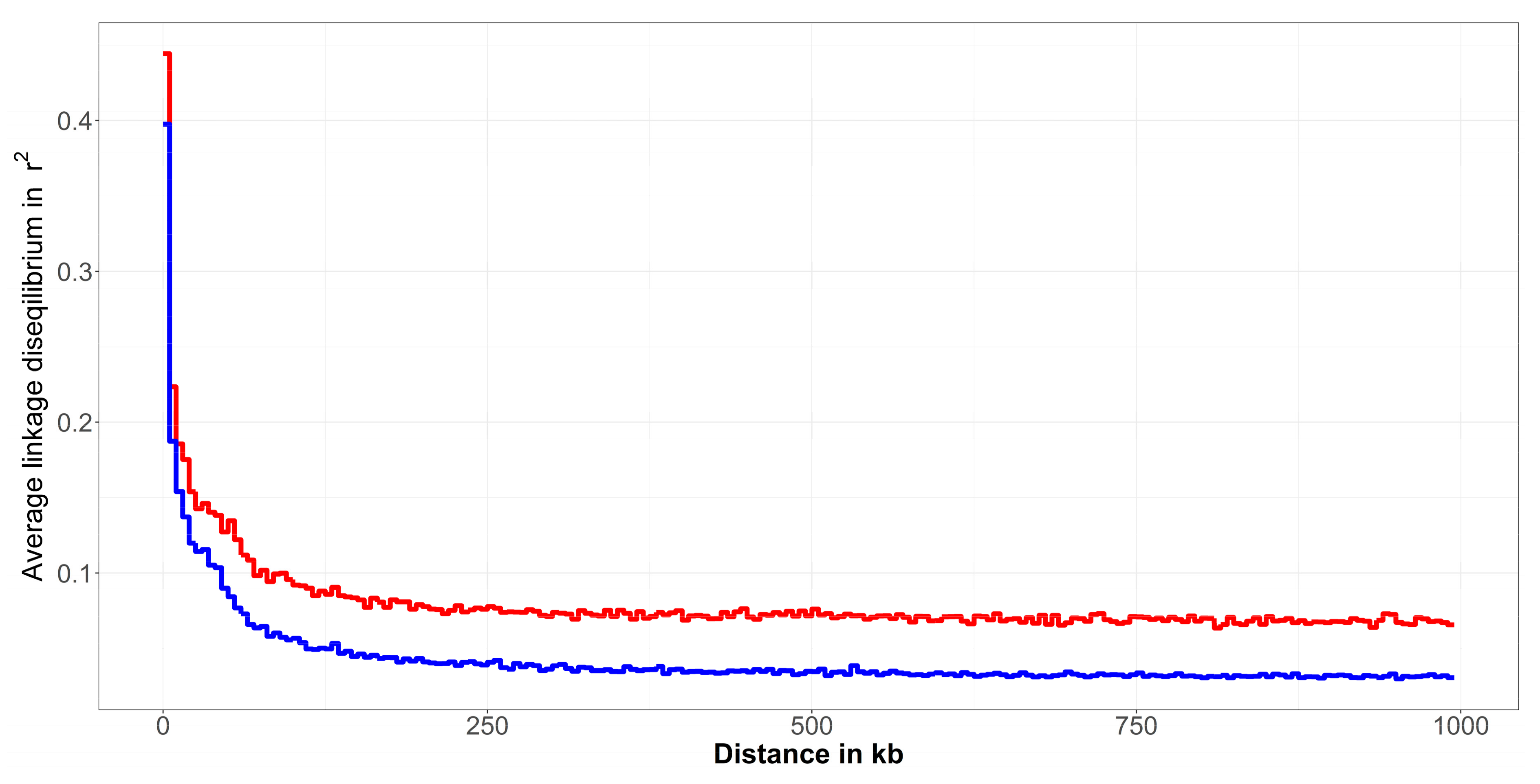
3.3. Linkage Disequilibrium
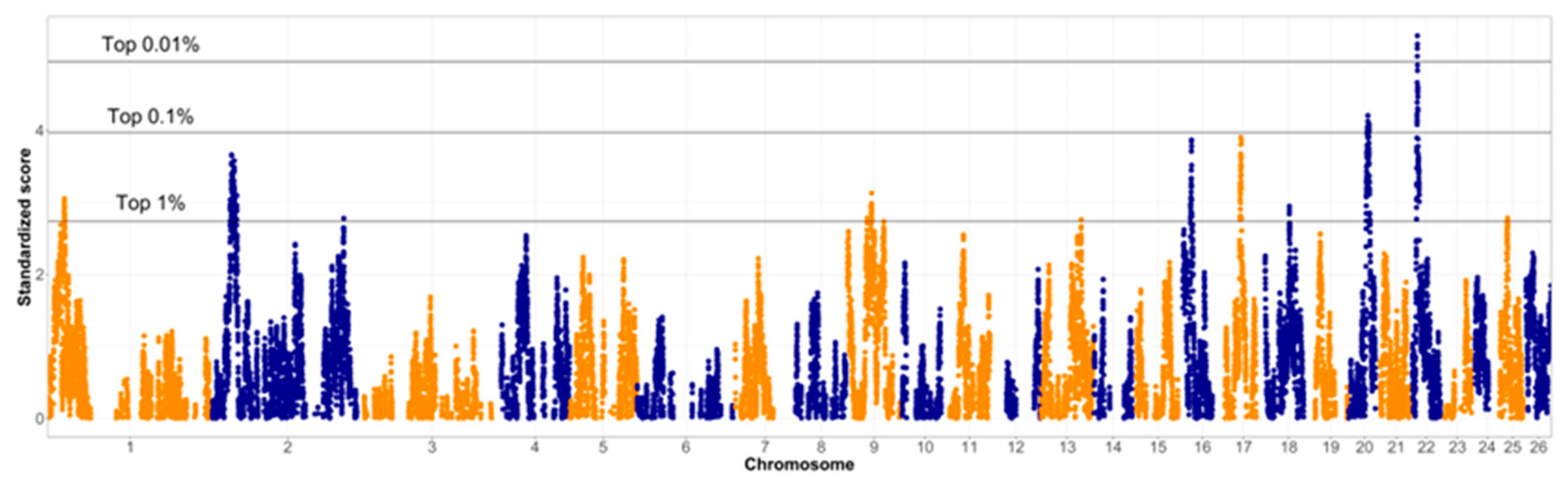
3.4. Between-Breed Selection Signatures Based on Variation of LD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeder, M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA 2008, 105, 11597–11604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A Genome Wide Survey of SNP Variation Reveals the Genetic Structure of Sheep Breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Neto, L.R.P.; Cristobal, M.S.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherf, B.D. World Watch List for Domestic Animal Diversity; Food and Agriculture Organization (FAO): Rome, Italy, 2000; Available online: https://www.cabdirect.org/cabdirect/abstract/20013016397 (accessed on 28 November 2021).
- Charlesworth, B.; Harvey, P.H.; Barton, N.H. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1553–1562. [Google Scholar] [CrossRef]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; Bi.Ov. Ita Consortium. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Mastrangelo, S.; Tolone, M.; Sardina, M.T.; Sottile, G.; Sutera, A.M.; Di Gerlando, R.; Portolano, B. Genome-wide scan for runs of homozygosity identifies potential candidate genes associated with local adaptation in Valle del Belice sheep. Genet. Sel. Evol. 2017, 49, 84. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Shi, L.; Li, Y.; Chen, L.; Garrick, D.; Wang, L.; Zhao, F. Estimates of genomic inbreeding and identification of candidate regions that differ between Chinese indigenous sheep breeds. J. Anim. Sci. Biotechnol. 2021, 12, 95. [Google Scholar] [CrossRef]
- Dzomba, E.F.; Chimonyo, M.; Pierneef, R.; Muchadeyi, F.C. Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genom. 2021, 22, 7. [Google Scholar] [CrossRef]
- Pérez O’Brien, A.M.; Utsunomiya, Y.T.; Mészáros, G.; Bickhart, D.M.; Liu, G.E.; Van Tassell, C.P.; Sonstegard, T.S.; Da Silva, M.V.; Garcia, J.F.; Sölkner, J. Assessing signatures of selection through variation in linkage disequilibrium between taurine and indicine cattle. Genet. Sel. Evol. 2014, 46, 19. [Google Scholar] [CrossRef] [Green Version]
- Sorbolini, S.; Marras, G.; Gaspa, G.; Dimauro, C.; Cellesi, M.; Valentini, A.; Macciotta, N.P. Detection of selection signatures in Piemontese and Marchigiana cattle, two breeds with similar production aptitudes but different selection histories. Genet. Sel. Evol. 2015, 47, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kukučková, V.; Moravčíková, N.; Kasarda, R. Evaluating signatures of selection through variation in linkage disequilibrium among different cattle breeds. Acta Fytotech. Zootech. 2016, 19, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, S.; Dong, K.; Tang, Z.; Li, K.; Fan, B.; Wang, Z.; Liu, B. Identification of positive selection signatures in pigs by comparing linkage disequilibrium variances. Anim. Genet. 2017, 48, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; McGivney, B.A.; Farries, G.; Katz, L.M.; MacHugh, D.E.; Randhawa, I.A.S.; Hill, E.W. Selection in Australian Thoroughbred horses acts on a locus associated with early two-year old speed. PLoS ONE 2020, 15, e0227212. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Zhang, L.; Orloff, M.S.; Reber, S.; Li, S.; Zhao, Y.; Eng, C. cgaTOH: Extended Approach for Identifying Tracts of Homozygosity. PLoS ONE 2013, 8, e57772. [Google Scholar] [CrossRef]
- Ferenčaković, M.; Sölkner, J.; Curik, I. Estimating autozygosity from high-throughput information: Effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013, 45, 42. [Google Scholar] [CrossRef] [Green Version]
- Ong, R.T.-H.; Teo, Y.-Y. varLD: A program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics 2010, 26, 1269–1270. [Google Scholar] [CrossRef]
- Arnaboldi, L.; Ossoli, A.; Giorgio, E.; Pisciotta, L.; Lucchi, T.; Grigore, L.; Pavanello, C.; Granata, A.; Pasta, A.; Arosio, B.; et al. LIPA gene mutations affect the composition of lipoproteins: Enrichment in ACAT-derived cholesteryl esters. Atherosclerosis 2020, 297, 8–15. [Google Scholar] [CrossRef]
- Deng, X.; Wang, D.; Wang, S.; Wang, H.; Zhou, H. Identification of key genes and pathways involved in response to pain in goat and sheep by transcriptome sequencing. Biol. Res. 2018, 51, 25. [Google Scholar] [CrossRef]
- Tang, J.; Hu, W.; Chen, S.; Di, R.; Liu, Q.; Wang, X.; He, X.; Gan, S.; Zhang, X.; Zhang, J.; et al. The genetic mechanism of high prolificacy in small tail han sheep by comparative proteomics of ovaries in the follicular and luteal stages. J. Proteom. 2019, 204, 103394. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, F.; Hunt, P.; Li, C.; Zhang, L.; Ingham, A.; Li, R.W. Transcriptome analysis unraveled potential mechanisms of resistance to Haemonchus contortus infection in Merino sheep populations bred for parasite resistance. Vet. Res. 2019, 50, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonczyk, M.S.; Simon, M.; Kumar, S.; Fernandes, V.E.; Sylvius, N.; Mallon, A.-M.; Denny, P.; Andrew, P.W. Genetic Factors Regulating Lung Vasculature and Immune Cell Functions Associate with Resistance to Pneumococcal Infection. PLoS ONE 2014, 9, e89831. [Google Scholar] [CrossRef]
- Mousel, M.R.; White, S.N.; Herndon, M.K.; Herndon, D.R.; Taylor, J.B.; Becker, G.M.; Murdoch, B.M. Genes involved in immune, gene translation and chromatin organization pathways associated with Mycoplasma ovipneumoniae presence in nasal secretions of domestic sheep. PLoS ONE 2021, 16, e0247209. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Mele, V.; Liberatore, R.A.; Bieniasz, P.D. Inhibition of spumavirus gene expression by PHF11. PLoS Pathog. 2020, 16, e1008644. [Google Scholar] [CrossRef]
- Rahman, N.; Stewart, G.; Jones, G. A role for the atopy-associated gene PHF11 in T-cell activation and viability. Immunol. Cell Biol. 2010, 88, 817–824. [Google Scholar] [CrossRef]
- Crameri, A.; Biondi, E.; Kuehnle, K.; Lütjohann, D.; Thelen, K.M.; Perga, S.; Dotti, C.G.; Nitsch, R.M.; Ledesma, M.D.; Mohajeri, M.H. The role of seladin-1/DHCR24 in cholesterol biosynthesis, APP processing and Abeta generation in vivo. EMBO J. 2006, 25, 432–443. [Google Scholar] [CrossRef] [Green Version]
- Ilie, D.E.; Mizeranschi, A.E.; Mihali, C.V.; Neamț, R.I.; Goilean, G.V.; Georgescu, O.I.; Zaharie, D.; Carabaș, M.; Huțu, I. Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes 2021, 12, 1495. [Google Scholar] [CrossRef]
- Granner, D.; Pilkis, S. The genes of hepatic glucose metabolism. J. Biol. Chem. 1990, 265, 10173–10176. [Google Scholar] [CrossRef]
- Vineeth, M.R.; Surya, T.; Sivalingam, J.; Kumar, A.; Niranjan, S.K.; Dixit, S.P.; Singh, K.; Tantia, M.S.; Gupta, I.D. Genome-wide discovery of SNPs in candidate genes related to production and fertility traits in Sahiwal cattle. Trop. Anim. Health Prod. 2020, 52, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Goldammer, T.; Brunner, R.M.; Kuehn, C. Tissue-specific mRNA expression patterns reveal a coordinated metabolic response associated with genetic selection for milk production in cows. Physiol. Genom. 2012, 44, 728–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medrano, J.F.; Sharrow, L. Milk Protein Typing of Bovine Mammary Gland Tissue Used to Generate a Complementary Deoxyribonucleic Acid Library. J. Dairy Sci. 1989, 72, 3190–3196. [Google Scholar] [CrossRef]
- Baba, N.A.; Panigrahi, M.; Verma, A.D.; Sadam, A.; Sulabh, S.; Chhotaray, S.; Parida, S.; Krishnaswamy, N.; Bhushan, B. Endometrial transcript profile of progesterone-regulated genes during early pregnancy of Water Buffalo (Bubalus bubalis). Reprod. Domest. Anim. 2019, 54, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, A.; Hebbar, M.; Srivastava, A.; Kadavigere, R.; Upadhyai, P.; Kanthi, A.; Brandau, O.; Bielas, S.; Girisha, K.M. Homozygous p.(Glu87Lys) variant in ISCA1 is associated with a multiple mitochondrial dysfunctions syndrome. J. Hum. Genet. 2017, 62, 723–727. [Google Scholar] [CrossRef] [Green Version]
- Esmaeili-Fard, S.M.; Gholizadeh, M.; Hafezian, S.H.; Abdollahi-Arpanahi, R. Genome-wide association study and pathway analysis identify NTRK2 as a novel candidate gene for litter size in sheep. PLoS ONE 2021, 16, e0244408. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Dutt, T.; Mishra, B.P. Genome-wide analysis of genetic diversity and selection signatures in three Indian sheep breeds. Livest. Sci. 2021, 243, 104367. [Google Scholar] [CrossRef]
- Parker Gaddis, K.L.; Null, D.J.; Cole, J.B. Explorations in genome-wide association studies and network analyses with dairy cattle fertility traits. J. Dairy Sci. 2016, 99, 6420–6435. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, J.; Zheng, H.; Xu, C.; Yan, W. UBQLN1 interacts with SPEM1 and participates in spermiogenesis. Mol. Cell. Endocrinol. 2010, 327, 89. [Google Scholar] [CrossRef] [Green Version]
- Østrup, E.; Hyttel, P.; Østrup, O.; Østrup, E.; Hyttel, P.; Østrup, O. Embryo–maternal communication: Signalling before and during placentation in cattle and pig. Reprod. Fertil. Dev. 2011, 23, 964–975. [Google Scholar] [CrossRef]
- Tidwell, J.A.; Schmidt, C.; Heaton, P.; Wilson, V.; Tucker, P.W. Characterization of a new ARID family transcription factor (Brightlike/ARID3C) that co-activates Bright/ARID3A-mediated immunoglobulin gene transcription. Mol. Immunol. 2011, 49, 260–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Peñagaricano, F.; Weigel, K.A.; Zhang, Y.; Rosa, G.; Khatib, H. Comparative genomics between fly, mouse, and cattle identifies genes associated with sire conception rate. J. Dairy Sci. 2012, 95, 6122–6129. [Google Scholar] [CrossRef] [PubMed]
- Serranito, B.; Cavalazzi, M.; Vidal, P.; Taurisson-Mouret, D.; Ciani, E.; Bal, M.; Rouvellac, E.; Servin, B.; Moreno-Romieux, C.; Tosser-Klopp, G.; et al. Local adaptations of Mediterranean sheep and goats through an integrative approach. Sci. Rep. 2021, 11, 21363. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.; Danner, A.L.; Famula, T.R.; Oberbauer, A.M. Genome-Wide Association Studies Reveal Susceptibility Loci for Noninfectious Claw Lesions in Holstein Dairy Cattle. Front. Genet. 2021, 12, 657375. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Eriksson, S.; Johansson, A.M.; Khansefid, M.; Moghaddaszadeh-Ahrabi, S.; Parna, N.; Davoudi, P.; Javanmard, A. Signatures of selection reveal candidate genes involved in economic traits and cold acclimation in five Swedish cattle breeds. Genet. Sel. Evol. 2020, 52, 52. [Google Scholar] [CrossRef]
- Valsalan, J.; Sadan, T.; Venkatachalapathy, T.; Anilkumar, K.; Aravindakshan, T.V. Identification of novel single-nucleotide polymorphism at exon1 and 2 region of B4GALT1 gene and its association with milk production traits in crossbred cattle of Kerala, India. Anim. Biotechnol. 2021, 1–9. [Google Scholar] [CrossRef]
- Banos, G.; Clark, E.L.; Bush, S.J.; Dutta, P.; Bramis, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. Genomic analyses underpin the feasibility of concomitant genetic improvement of milk yield and mastitis resistance in dairy sheep. PLoS ONE 2019, 14, e0214346. [Google Scholar] [CrossRef] [Green Version]
- Malheiros, J.M.; Enríquez-Valencia, C.E.; da Silva Duran, B.O.; de Paula, T.G.; Curi, R.A.; de Vasconcelos Silva, J.A.I.I.; Dal-Pai-Silva, M.; de Oliveira, H.N.; Chardulo, L.A.L. Association of CAST2, HSP90AA1, DNAJA1 and HSPB1 genes with meat tenderness in Nellore cattle. Meat Sci. 2018, 138, 49–52. [Google Scholar] [CrossRef]
- Lim, D.; Lee, S.-H.; Kim, N.-K.; Cho, Y.-M.; Chai, H.-H.; Seong, H.-H.; Kim, H. Gene Co-expression Analysis to Characterize Genes Related to Marbling Trait in Hanwoo (Korean) Cattle. Asian-Aust. J. Anim. Sci. 2013, 26, 19–29. [Google Scholar] [CrossRef]
- Yang, F.; Chen, F.; Li, L.; Yan, L.; Badri, T.; Lv, C.; Yu, D.; Zhang, M.; Jang, X.; Li, J.; et al. Three Novel Players: PTK2B, SYK, and TNFRSF21 Were Identified to Be Involved in the Regulation of Bovine Mastitis Susceptibility via GWAS and Post-transcriptional Analysis. Front. Immunol. 2019, 10, 1579. [Google Scholar] [CrossRef] [Green Version]
- Khanna, R.; Wilson, S.M.; Brittain, J.M.; Weimer, J.; Sultana, R.; Butterfield, A.; Hensley, K. Opening Pandora’s jar: A primer on the putative roles of CRMP2 in a panoply of neurodegenerative, sensory and motor neuron, and central disorders. Future Neurol. 2012, 7, 749–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posbergh, C.J.; Thonney, M.L.; Huson, H.J. Genomic Approaches Identify Novel Gene Associations with Out of Season Lambing in Sheep. J. Hered. 2019, 110, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Stine, R.R.; Shapira, S.N.; Lim, H.-W.; Ishibashi, J.; Harms, M.; Won, K.-J.; Seale, P. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab. 2016, 5, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, P.; Amorín, R.; Han, Y.; Peñagaricano, F. Whole-genome scan reveals significant non-additive effects for sire conception rate in Holstein cattle. BMC Genet. 2018, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Song, G.; Kim, J.; Dunlap, K.A.; Satterfield, M.C.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Uterine biology in pigs and sheep. J. Anim. Sci. Biotechnol. 2012, 3, 23. [Google Scholar] [CrossRef] [Green Version]
- Connor, E.E.; Siferd, S.; Elsasser, T.H.; Evock-Clover, C.M.; Van Tassell, C.P.; Sonstegard, T.S.; Fernandes, V.M.; Capuco, A.V. Effects of increased milking frequency on gene expression in the bovine mammary gland. BMC Genom. 2008, 9, 362. [Google Scholar] [CrossRef]
- Abdollahi-Arpanahi, R.; Pacheco, H.A.; Peñagaricano, F. Targeted sequencing reveals candidate causal variants for dairy bull subfertility. Anim. Genet. 2021, 52, 509–513. [Google Scholar] [CrossRef]
- Kour, A.; Deb, S.M.; Nayee, N.; Niranjan, S.K.; Raina, V.S.; Mukherjee, A.; Gupta, I.D.; Patil, C.S. Novel insights into genome-wide associations in Bos indicus reveal genetic linkages between fertility and growth. Anim. Biotechnol. 2021, 1–17. [Google Scholar] [CrossRef]
- Mohammadi, H.; Rafat, S.A.; Moradi Shahrbabak, H.; Shodja, J.; Moradi, M.H. Genome-wide association study and gene ontology for growth and wool characteristics in Zandi sheep. J. Livest. Sci. Technol. 2020, 8, 45–55. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide scan of selection signatures in Dehong humped cattle for heat tolerance and disease resistance. Anim. Genet. 2020, 51, 292–299. [Google Scholar] [CrossRef]
- Fischer, D.; Laiho, A.; Gyenesei, A.; Sironen, A. Identification of Reproduction-Related Gene Polymorphisms Using Whole Transcriptome Sequencing in the Large White Pig Population. G3 Genes Genomes Genet. 2015, 5, 1351–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Zhu, Y.; Ding, Y.; Huang, Z.; Dan, X.; Shi, Y.; Kang, X. Identifying the key genes and functional enrichment pathways associated with feed efficiency in cattle. Gene 2022, 807, 145934. [Google Scholar] [CrossRef] [PubMed]
- Marín-Garzón, N.A.; Magalhães, A.F.B.; Mota, L.F.M.; Fonseca, L.F.S.; Chardulo, L.A.L.; Albuquerque, L.G. Genome-wide association study identified genomic regions and putative candidate genes affecting meat color traits in Nellore cattle. Meat Sci. 2021, 171, 108288. [Google Scholar] [CrossRef]
- Sakai, K.; Ito, C.; Wakabayashi, M.; Kanzaki, S.; Ito, T.; Takada, S.; Toshimori, K.; Sekita, Y.; Kimura, T. Usp26 mutation in mice leads to defective spermatogenesis depending on genetic background. Sci. Rep. 2019, 9, 13757. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-T.; Chung, W.-H.; Lee, S.-Y.; Choi, J.-W.; Kim, J.; Lim, D.; Lee, S.; Jang, G.-W.; Kim, B.; Choy, Y.H.; et al. Whole-genome resequencing of Hanwoo (Korean cattle) and insight into regions of homozygosity. BMC Genom. 2013, 14, 519. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhu, J.; He, Q.; Loor, J.J.; Luo, J. Association between the expression of miR-26 and goat milk fatty acids. Reprod. Domest. Anim. 2018, 53, 1478–1482. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Ganguly, I.; Singh, R.; Deb, S.M.; Kumar, S.; Sharma, A.; Mitra, A. DNA Polymorphism in SLC11A1 Gene and its Association with Brucellosis Resistance in Indian Zebu (Bos indicus) and Crossbred (Bos indicus×Bos taurus) Cattle. Asian-Aust. J. Anim. Sci. 2011, 24, 898–904. [Google Scholar] [CrossRef]
- Taka, S.; Liandris, E.; Gazouli, M.; Sotirakoglou, K.; Theodoropoulos, G.; Bountouri, M.; Andreadou, M.; Ikonomopoulos, J. In vitro expression of the SLC11A1 gene in goat monocyte-derived macrophages challenged with Mycobacterium avium subsp paratuberculosis. Infect. Genet. Evol. 2013, 17, 8–15. [Google Scholar] [CrossRef]
- Wu, H.; Pan, Y.; Zhang, Q.; Cao, Y.; Li, J.; Chen, H.; Cai, Y.; Sun, X.; Lan, X. Insertion/deletion (InDel) variations in sheep PLAG1 gene locating in growth-related major QTL are associated with adult body weight and morphometric traits. Small Rumin. Res. 2019, 178, 63–69. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, M.; Wu, H.; Akhatayeva, Z.; Lan, X.; Fei, P.; Mao, C.; Jiang, F. Indel mutations of sheep PLAG1 gene and their associations with growth traits. Anim. Biotechnol. 2021, 1–7. [Google Scholar] [CrossRef]
- Taye, M.; Yoon, J.; Dessie, T.; Cho, S.; Oh, S.J.; Lee, H.-K.; Kim, H. Deciphering signature of selection affecting beef quality traits in Angus cattle. Genes Genom. 2018, 40, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Shen, D.; Li, C.; Cai, W.; Liu, S.; Yin, H.; Shi, S.; Cao, M.; Zhang, S. Comparative Transcriptomic and Proteomic Analyses Identify Key Genes Associated With Milk Fat Traits in Chinese Holstein Cows. Front. Genet. 2019, 10, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vohra, V.; Chhotaray, S.; Gowane, G.; Alex, R.; Mukherjee, A.; Verma, A.; Deb, S.M. Genome-Wide Association Studies in Indian Buffalo Revealed Genomic Regions for Lactation and Fertility. Front. Genet. 2021, 12, 1668. [Google Scholar] [CrossRef] [PubMed]
- Fleming-Waddell, J.N.; Olbricht, G.R.; Taxis, T.M.; White, J.D.; Vuocolo, T.; Craig, B.A.; Tellam, R.L.; Neary, M.K.; Cockett, N.E.; Bidwell, C.A. Effect of DLK1 and RTL1 but Not MEG3 or MEG8 on Muscle Gene Expression in Callipyge Lambs. PLoS ONE 2009, 4, e7399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, C.; Cassar-Malek, I.; Le Cunff, M.; Dubroeucq, H.; Renand, G.; Hocquette, J.-F. New Indicators of Beef Sensory Quality Revealed by Expression of Specific Genes. J. Agric. Food Chem. 2007, 55, 5229–5237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Zhang, B.; Wang, Z.; Zhang, L.; Chen, H.; Zhou, D.; Tang, K.; Wang, A.; Lin, P.; Jin, Y. COPS5 negatively regulates goat endometrial function via the ERN1 and mTOR-autophagy pathways during early pregnancy. J. Cell. Physiol. 2019, 234, 18666–18678. [Google Scholar] [CrossRef]
- Sun, W.; Hudson, N.J.; Reverter, A.; Waardenberg, A.J.; Tellam, R.L.; Vuocolo, T.; Byrne, K.; Dalrymple, B.P. An Always Correlated gene expression landscape for ovine skeletal muscle, lessons learnt from comparison with an “equivalent” bovine landscape. BMC Res. Notes 2012, 5, 632. [Google Scholar] [CrossRef] [Green Version]
- Ramayo-Caldas, Y.; Fortes, M.R.S.; Hudson, N.J.; Porto-Neto, L.R.; Bolormaa, S.; Barendse, W.; Kelly, M.; Moore, S.S.; Goddard, M.E.; Lehnert, S.A.; et al. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J. Anim. Sci. 2014, 92, 2832–2845. [Google Scholar] [CrossRef]
- Naserkheil, M.; Bahrami, A.; Lee, D.; Mehrban, H. Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle. Animals 2020, 10, 1836. [Google Scholar] [CrossRef]
- Bai, M.; Sun, L.; Zhao, J.; Xiang, L.; Cheng, X.; Li, J.; Jia, C.; Jiang, H. Histological analysis and identification of spermatogenesis-related genes in 2-, 6-, and 12-month-old sheep testes. Sci. Nat. 2017, 104, 84. [Google Scholar] [CrossRef]
- Intergenomics Consortium; Frischknecht, M.; Bapst, B.; Seefried, F.R.; Signer-Hasler, H.; Garrick, D.; Stricker, C.; Fries, R.; Russ, I.; Sölkner, J.; et al. Genome-wide association studies of fertility and calving traits in Brown Swiss cattle using imputed whole-genome sequences. BMC Genom. 2017, 18, 910. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Mipam, T.; Xu, C.; Zhao, W.; Shah, M.A.; Yi, C.; Luo, H.; Cai, X.; Zhong, J. Testis transcriptome profiling identified genes involved in spermatogenic arrest of cattleyak. PLoS ONE 2020, 15, e0229503. [Google Scholar] [CrossRef] [PubMed]
- Sharp, J.A.; Lefèvre, C.; Watt, A.; Nicholas, K.R. Analysis of human breast milk cells: Gene expression profiles during pregnancy, lactation, involution, and mastitic infection. Funct. Integr. Genom. 2016, 16, 297–321. [Google Scholar] [CrossRef] [PubMed]
- Mömke, S.; Sickinger, M.; Lichtner, P.; Doll, K.; Rehage, J.; Distl, O. Genome-wide association analysis identifies loci for left-sided displacement of the abomasum in German Holstein cattle. J. Dairy Sci. 2013, 96, 3959–3964. [Google Scholar] [CrossRef] [Green Version]
- Buitenhuis, B.; Janss, L.L.; Poulsen, N.A.; Larsen, L.B.; Larsen, M.K.; Sørensen, P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom. 2014, 15, 1112. [Google Scholar] [CrossRef] [Green Version]
- Weedon, M.N.; Lango, H.; Lindgren, C.M.; Wallace, C.; Evans, D.M.; Mangino, M.; Freathy, R.M.; Perry, J.R.B.; Stevens, S.; Hall, A.S.; et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008, 40, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Z.; Medland, S.E.; Wright, M.J.; Heath, A.C.; Madden, P.A.F.; Duncan, A.; Montgomery, G.W.; Martin, N.G.; McRae, A.F. Genome-wide association study of height and body mass index in Australian twin families. Twin Res. Hum. Genet. Off. J. Int. Soc. Twin Stud. 2010, 13, 179. [Google Scholar] [CrossRef]
- Nonneman, D.J.; Schneider, J.F.; Lents, C.A.; Wiedmann, R.T.; Vallet, J.L.; Rohrer, G.A. Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 2016, 17, 50. [Google Scholar] [CrossRef] [Green Version]
- Vuocolo, T.; Byrne, K.; White, J.; McWilliam, S.; Reverter, A.; Cockett, N.E.; Tellam, R.L. Identification of a gene network contributing to hypertrophy in callipyge skeletal muscle. Physiol. Genom. 2007, 28, 253–272. [Google Scholar] [CrossRef] [Green Version]

| FROH (Up to 3 Generations) | FROH (Up to 6 Generations) | FROH (Up to 12 Generations) | FROH (Up to 25 Generations) | FROH (Up to 50 Generations) | |
|---|---|---|---|---|---|
| OV | 0.020 ± 0.026 | 0.038 ± 0.040 | 0.058 ± 0.052 | 0.068 ± 0.058 | 0.079 ± 0.064 |
| IV | 0.001 ± 0.006 | 0.003 ± 0.007 | 0.006 ± 0.010 | 0.008 ± 0.010 | 0.013 ± 0.011 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mészárosová, M.; Mészáros, G.; Moravčíková, N.; Pavlík, I.; Margetín, M.; Kasarda, R. Within- and between-Breed Selection Signatures in the Original and Improved Valachian Sheep. Animals 2022, 12, 1346. https://doi.org/10.3390/ani12111346
Mészárosová M, Mészáros G, Moravčíková N, Pavlík I, Margetín M, Kasarda R. Within- and between-Breed Selection Signatures in the Original and Improved Valachian Sheep. Animals. 2022; 12(11):1346. https://doi.org/10.3390/ani12111346
Chicago/Turabian StyleMészárosová, Mária, Gábor Mészáros, Nina Moravčíková, Ivan Pavlík, Milan Margetín, and Radovan Kasarda. 2022. "Within- and between-Breed Selection Signatures in the Original and Improved Valachian Sheep" Animals 12, no. 11: 1346. https://doi.org/10.3390/ani12111346






