Analysis of the Genetic Diversity and Population Structure of Four Senegalese Sheep Breeds Using Medium-Density Single-Nucleotide Polymorphisms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Breeds, Sampling Strategy, and DNA Extraction
2.2. DNA Genotyping
2.3. Analyses of Sheep Breed Genetic Diversity and Structure
3. Results and Discussion
3.1. Within-Breed Genetic Diversity
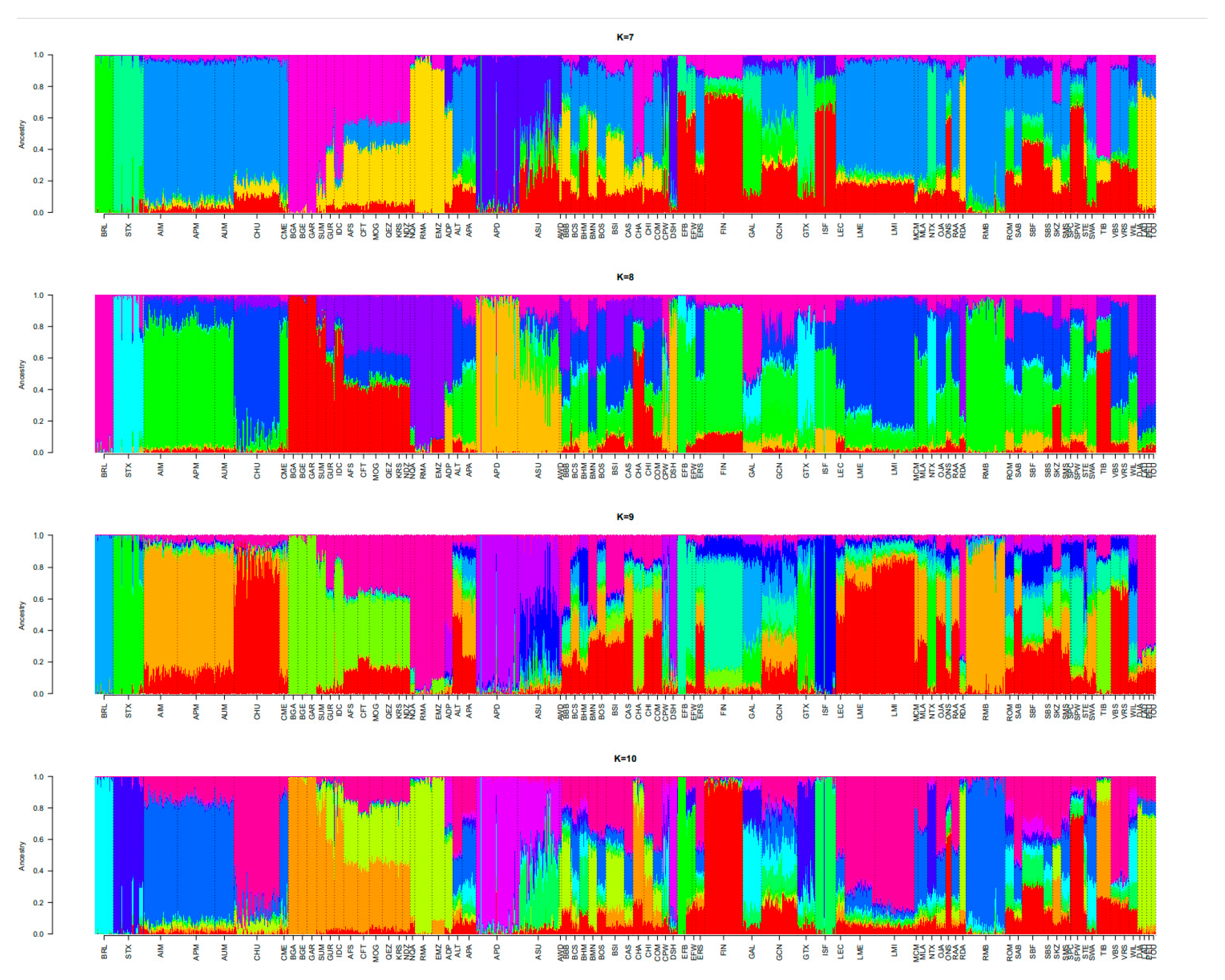
3.2. Population Relationships and Structure
3.3. Senegalese Sheep Breeds in the Global Context
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Seydi, M.; Ba, Y.M. Les viandes rechercheés par les Sénégalais. Viandes Et Prod. Carnés 1992, 13, 139–142. [Google Scholar]
- Ninot, O. Des moutons pour la fête: L’approvisionnement de Dakar en moutons de Tabaski. Les Cah. D’outre-Mer 2010, 249, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Brisebarre, A.M. La Fête de la Tabaski en Milieu Urbain au Sénégal. Enjeux Culturels, Sociaux et Économiques; Rapport de Recherche; Conseil pour le Développement de la Recherche en Sciences Sociales en Afrique (CODESRIA); Institut de Recherche pour le Développement (IRD): Dakar, Senegal, 2003; p. 402. [Google Scholar]
- Bruford, M.W.; Townsend, S.J. Mitochondridal DNA diversity in modern sheep. In Documenting Domestication: New Genetic and Archaeological Paradigms; Zeder, M.A., Bradley, D.G., Emshwiller, E., Smith, B.D., Eds.; University of California Press: Berkeley/Los Angeles, CA, USA, 2006; pp. 306–316. [Google Scholar]
- Close, A. Holocene occupation of the eastern Sahara. In New Light on the Northeast African Past; Klees, F., Kuper, R., Eds.; Heinrich-Barth-Institut: Köln, Germany, 1992; pp. 157–183. [Google Scholar]
- Ryder, M.L. Sheep and Man; Duckworth: London, UK, 1983. [Google Scholar]
- Ryder, M.L. Sheep. In Evolution of Domesticated Animals; Mason, I.L., Ed.; Longman: London, UK, 1984; pp. 63–85. [Google Scholar]
- Fall, A. Peul, Touabire and Djallonke sheep breeding programmes in Senegal. In Proceedings of the Workshop on Developing Breeding Strategies for Lower Input Animal Production Environments, Bella, Italy, 22–25 September 1999; International Committee for Animal Recording: Rome, Italy, 2000; pp. 331–338. [Google Scholar]
- Ndiaye, B.; Diouf, M.N.; Ciss, M.; Wane, M.; Diop, M.; Sembène, M. Morphologie et pratiques d’élevage du mouton peul-peul du sénégal. Int. J. Adv. Res. 2018, 6, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Ousseini, H. Analyse Socioéconomique des Élevages du Mouton Ladoum Dans la Commune de Thiès/Sénégal. Master’s Thesis, EISMV. Université Cheikh Anta Diop, Dakar, Senegal, 2011. [Google Scholar]
- Edea, Z.; Dessie, T.; Dadi, H.; Do, K.-T.; Kim, K.-S. Genetic Diversity and Population Structure of Ethiopian Sheep Populations Revealed by High-Density SNP Markers. Front. Genet. 2017, 8, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gueye, A. Moutons et Chèvres du Sénégal: Caractérisation Morphobiométrique et Typage Sanguin. Ph.D. Thesis, Université Cheikh Anta Diop, Dakar, Senegal, 1997. [Google Scholar]
- Missohou, A.; Nguyen, T.C.; Sow, R.; Gueye, A. Blood Polymorphism in West African Breeds of Sheep. Trop. Anim. Health Prod. 1999, 31, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A Genome Wide Survey of SNP Variation Reveals the Genetic Structure of Sheep Breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, W.-R.; Lv, F.-H.; He, S.-G.; Tian, S.-L.; Peng, W.-F.; Sun, Y.-W.; Zhao, Y.-X.; Tu, X.-L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.X.; Yang, J.; Lv, F.H.; Hu, X.J.; Xie, X.L.; Zhang, M.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef]
- Sow, R.S.; Thiongane, P.I.; Tchamutchina, L. Bilan de cinq années d’études des moutons Peul et Touabire au Centre de Recherche Zootechnique de Dahra-Djoloff. Rev. Sénégalaise Des. Rech. Agric. Et Halieut. 1988, 1, 80–89. [Google Scholar]
- Jeanpierre, M. A rapid method for the purification of DNA from blood. Nucleic Acids Res 1987, 15, 9611. [Google Scholar] [CrossRef] [Green Version]
- Sempéré, G.; Moazami-Goudarzi, K.; Eggen, A.; Laloë, D.; Gautier, M.; Flori, L. WIDDE: A Web-Interfaced next generation database for genetic diversity exploration, with a first application in cattle. BMC Genom. 2015, 16, 940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciani, E.; Lasagna, E.; D’Andrea, M.; Alloggio, I.; Marroni, F.; Ceccobelli, S.; Delgado Bermejo, J.V.; Sarti, F.M.; Kijas, J.; Lenstra, J.A.; et al. Merino and Merino-derived sheep breeds: A genome-wide intercontinental study. Genet. Sel. Evol. 2015, 47, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, E.A.; Kijas, J.W.; Mcculloch, R.; Scobie, D.R.; Mcrae, K.M.; Pickering, N.K.; Dodds, K.G.; Mcewan, J.C. Arapawa: A novel New Zealand sheep breed of distinct origin. Proc. New Zealand Soc. Anim. Prod. 2011, 71, 248–250. [Google Scholar]
- Patterson, N.; Price, A.L.; Reich, D. Population Structure and Eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Chessel, D.; Dufour, A.; Thioulouse, J. The ade4 package-I-One-table methods. R News 2004, 4, 5–10. [Google Scholar]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- De Meeûs, T. Initiation à la Génétique des Populations Naturelles: Application aux Parasites et à Leurs Vecteurs; IRD Editions: Marseilles, France, 2012. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [Green Version]
- Beynon, S.E.; Slavov, G.T.; Farré, M.; Sunduimijid, B.; Waddams, K.; Davies, B.; Haresign, W.; Kijas, J.; MacLeod, I.M.; Newbold, C.J.; et al. Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet. 2015, 16, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molotsi, A.H.; Taylor, J.F.; Cloete, S.W.P.; Muchadeyi, F.; Decker, J.E.; Whitacre, L.K.; Sandenbergh, L.; Dzama, K. Genetic diversity and population structure of South African smallholder farmer sheep breeds determined using the OvineSNP50 beadchip. Trop. Anim. Health Prod. 2017, 49, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Spangler, G.L.; Rosen, B.D.; Ilori, M.B.; Hanotte, O.; Kim, E.-S.; Sonstegard, T.S.; Burke, J.M.; Morgan, J.L.M.; Notter, D.R.; Van Tassell, C.P. Whole genome structural analysis of Caribbean hair sheep reveals quantitative link to West African ancestry. PLoS ONE 2017, 12, e0179021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoda, A.; Hykaj, G.; Sena, L.; Delia, E. Population structure in three Albanian sheep breeds using 36 single nucleotide polymorphisms. Acta Agric. Scand. Sect. A Anim. Sci. 2011, 61, 12–20. [Google Scholar] [CrossRef]
- Álvarez, I.; Traoré, A.; Tamboura, H.H.; Kaboré, A.; Royo, L.J.; Fernández, I.; Ouédraogo-Sanou, G.; Sawadogo, L.; Goyache, F. Microsatellite Analysis Characterizes Burkina Faso as a Genetic Contact Zone Between Sahelian and Djallonké Sheep. Anim. Biotechnol. 2009, 20, 47–57. [Google Scholar] [CrossRef]
- Wafula, P.; Jianlin, H.; Sangare, N.; Sowe, J.; Coly, R.; Diallo, B.; Hanotte, O. Genetic characterization of West African djallonke sheep using microsatellite markers. Turin Role Biotechnol. 2005, 5, 47–57. [Google Scholar]
- Dayo, G.-K.; Houaga, I.; Somda, M.B.; Linguelegue, A.; Ira, M.; Konkobo, M.; Djassi, B.; Gomes, J.; Sangare, M.; Cassama, B.; et al. Morphological and microsatellite DNA diversity of Djallonké sheep in Guinea-Bissau. BMC Genom. Data 2022, 23, 3. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29. [Google Scholar] [CrossRef] [Green Version]
- MacHugh, D.E.; Shriver, M.D.; Loftus, R.T.; Cunningham, P.; Bradley, D.G. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 1997, 146, 1071–1086. [Google Scholar] [CrossRef]
- Missohou, A.; Adakal, E.H. Situation actuelle et perspectives d’une gestion durable des ressources génétiques bovines d’Afrique de l’Ouest. In Proceedings of the Colloque sur le Développement Durable: Leçons et Perspectives, Ouagadougou, Burkina Faso, 1–4 June 2004. [Google Scholar]
- Rege, J.E.O. The state of African cattle genetic resources I. Classification framework and identification of threatened and extinct breeds. Anim. Genet. Resour. Inf. 1999, 25, 1–25. [Google Scholar] [CrossRef]
- Cesaro, J.-D.; Magrin, G.; Minot, O. Atlas de L’élevage au Sénégal; Commerce et Territoires; CIRAD-ATP Icare/Université Paris I-Prodig: Paris, France, 2010; p. 32. [Google Scholar]
- Sandenbergh, L.; Cloete, S.W.P.; Roodt-Wilding, R.; Snyman, M.A.; Merwe, A.E.v.d. Genetic diversity and population structure of four South African sheep breeds. In Proceedings of the Association for the Advancement of Animal Breeding and Genetics, Lorne, Australia, 28–30 September 2015; pp. 294–297. [Google Scholar]
- Doutressoulle, G. L’élevage en Afrique Occidentale Française; Larose: Paris, France, 1947. [Google Scholar]
- Epstein, H. The Origin of the Domesticated Animals of Africa; Africana Publication Corporation: New York, NY, USA, 1971; Volume 2. [Google Scholar]
- Cañón, J.; García, D.; García-Atance, M.A.; Obexer-Ruff, G.; Lenstra, J.A.; Ajmone-Marsan, P.; Dunner, S. Geographical partitioning of goat diversity in Europe and the Middle East. Anim. Genet. 2006, 37, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G.; Erhardt, G.; Consortium, T.E. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim. Genet. 2007, 38, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Gifford-Gonzalez, D.; Hanotte, O. Domesticating Animals in Africa: Implications of Genetic and Archaeological Findings. J. World Prehistory 2011, 24, 1–23. [Google Scholar] [CrossRef] [Green Version]






| Breed ID | Breed Name | Location | Area of Origin | Nb | Reference |
|---|---|---|---|---|---|
| ADP | African Dorper | South Africa | Souh Africa | 21 | [20] |
| AFS | Afshari | Iran | Middle East | 37 | [20] |
| AIM | Australian Industry Merino | Australia | Europe | 88 | [20] |
| ALT | Altamurana | South Italia | Europe | 24 | [20] |
| APA | Arapawa | New Zealand | Europe | 37 | [20,22] |
| APD | Australian Poll Dorset | Australia | Europe | 108 | [20] |
| APM | Australian Poll Merino | Australia | Europe | 98 | [20] |
| ASU | Australian Suffolk | Australia | Europe | 109 | [20] |
| AUM | Australian Merino | Australia | Europe | 50 | [20] |
| AWD | African White Dorper | South Africa | South Africa | 6 | [20] |
| BBB | Barbados Black Belly | Barbados | Caribbean | 24 | [20] |
| BCS | Brazilian Creole | Brazil | America | 23 | [20] |
| BGA | Bangladeshi Garole | Bangladesh | Asia | 24 | [20] |
| BGE | Bangladeshi East BGE | Bangladesh | Asia | 24 | [20] |
| BHM | Black Headed Mountain | UK | Europe | 24 | [20] |
| BMN | Morada Nova | Brazil | America | 22 | [20] |
| BOS | Bundner Oberlander Sheep | Germany | Europe | 24 | [20] |
| BRL | Border Leicester | UK | Europe | 48 | [20] |
| BSI | Santa Ines | Brazil | America | 47 | [20] |
| CAS | Castellana | Spain | Europe | 23 | [20] |
| CFT | Cyprus Fat Tail | Cyprus | Middle East | 30 | [20] |
| CHA | Changthangi | Indian | Asia | 29 | [20] |
| CHI | Chios | Greece | Europe | 23 | [20] |
| CHU | Churra | Spain | Europe | 120 | [20] |
| CME | Chinese Merino | China | Europe | 23 | [20] |
| COM | Comisana | Italia | Europe | 24 | [20] |
| CPW | Australian Coopworth | Australia | Europe | 19 | [20] |
| DJA | Djallonke | Senegal | West Africa | 11 | this study |
| DSH | Dorset Horn | UK | Europe | 21 | [20] |
| EFB | East Friesian Brown | Germany | Europe | 39 | [20] |
| EFW | East Friesian White | Germany | Europe | 9 | [20] |
| EMZ | Ethiopian Menz | Ethiopia | East Africa | 34 | [20] |
| ERS | Engadine Red Sheep | Swiss | Europe | 24 | [20] |
| FIN | Finnsheep | Finland | Europe | 99 | [20] |
| GAL | Galway | UK | Europe | 49 | [20] |
| GAR | Indian Garole | India | Asia | 26 | [20] |
| GCN | Gulf Coast Native | US | America | 94 | [20] |
| GTX | German Texel | Germany | Europe | 46 | [20] |
| GUR | Garut | Indonesia | Asia | 22 | [20] |
| IDC | Deccani | India | Asia | 24 | [20] |
| ISF | Irish Suffolk | UK | Europe | 55 | [20] |
| KRS | Karakas | Turkey | Middle East | 18 | [20] |
| LAD | Ladoum | Senegal | West Africa | 12 | this study |
| LEC | Leccese | Italia | Europe | 24 | [20] |
| LME | Lacaune (meat) | France | Europe | 78 | [20] |
| LMI | Lacaune (milk) | France | Europe | 103 | [20] |
| MCM | Macarthur Merino | Australia | Europe | 10 | [20] |
| MLA | Merinolandschaf | Germany | Europe | 24 | [20] |
| MOG | Moghani | Iran | Middle East | 34 | [20] |
| NDZ | Norduz | Turkey | Middle East | 20 | [20] |
| NQA | Namaqua Afrikaner | South Africa | South Africa | 12 | [20] |
| NTX | New Zealand Texel | New Zealand | Europe | 24 | [20] |
| OJA | Ojalada | Spain | Europe | 24 | [20] |
| ONS | Old Norwegian Spaelsau | Norway | Europe | 15 | [20] |
| PEU | Peul | Senegal | West Africa | 12 | this study |
| QEZ | Qezel | Iran | Middle East | 35 | [20] |
| RAA | Rasa Aragonesa | Spain | Europe | 22 | [20] |
| RDA | Ronderib Afrikaner | South Africa | South Africa | 17 | [20] |
| RMA | Red Maasai | Kenya | East Africa | 45 | [20] |
| RMB | Merinos de Rambouillet | France | Europe | 102 | [20] |
| ROM | New Zealand Romney | New Zealand | Europe | 24 | [20] |
| SAB | Sardinian Ancestral Black | Italia | Europe | 20 | [20] |
| SBF | Scottish Blackface | UK | Europe | 56 | [20] |
| SBS | Swiss Black | Swiss | Europe | 24 | [20] |
| SKZ | Sakiz | Turkey | Middle East | 22 | [20] |
| SMS | Swiss Mirror | Swiss | Europe | 24 | [20] |
| SPC | Spael | Norway | Europe | 3 | [20] |
| SPW | Spael | Norway | Europe | 32 | [20] |
| STE | St Elizabeth | Jamaica | Caribbean | 10 | [20] |
| STX | ScottishTexel | UK | Europe | 80 | [20] |
| SUM | Sumatra | Indonesia | Asia | 24 | [20] |
| SWA | Swiss White Alpine | Switzerland | Europe | 24 | [20] |
| TIB | Tibetan | China | Asia | 37 | [20] |
| TOU | Touabire | Senegal | West Africa | 12 | this study |
| VBS | Valais Blacknose | Canada | Europe | 24 | [20] |
| VRS | Valais Red | Switzerland | Europe | 24 | [20] |
| WIL | Wiltshire | UK | Europe | 23 | [20] |
| Breed | Nb | Ho | He | Fis |
|---|---|---|---|---|
| Djallonke | 11 | 0.302 | 0.286 | 0.024 |
| Peul-peul | 12 | 0.344 | 0.322 | 0.034 |
| Touabire | 12 | 0.349 | 0.330 | 0.024 |
| Ladoum | 12 | 0.323 | 0.307 | 0.019 |
| Djallonke | Ladoum | Peul-Peul | Touabire | |
|---|---|---|---|---|
| Djallonke | - | 1.913 | 2.980 | 3.079 |
| Ladoum | 0.116 | - | 5.432 | 8.679 |
| Peul-peul | 0.077 | 0.044 | - | 73.279 |
| Touabire | 0.075 | 0.028 | 0.003 | - |
| Breed ID | Breed Name | Nb | Ho | He | Fis |
|---|---|---|---|---|---|
| ADP | African Dorper | 21 | 0.342 | 0.337 | −0.012 |
| AFS | Afshari | 37 | 0.355 | 0.348 | −0.020 |
| AIM | Australian Industry Merino | 88 | 0.367 | 0.374 | 0.019 |
| ALT | Altamurana | 24 | 0.360 | 0.369 | 0.026 |
| APA | Arapawa | 37 | 0.326 | 0.349 | 0.067 |
| APD | Australian Poll Dorset | 108 | 0.345 | 0.344 | −0.003 |
| APM | Australian Poll Merino | 98 | 0.372 | 0.375 | 0.008 |
| ASU | Australian Suffolk | 109 | 0.372 | 0.372 | −0.001 |
| AUM | Australian Merino | 50 | 0.363 | 0.374 | 0.031 |
| AWD | African White Dorper | 6 | 0.327 | 0.301 | −0.086 |
| BBB | Barbados Black Belly | 24 | 0.317 | 0.341 | 0.070 |
| BCS | Brazilian Creole | 23 | 0.329 | 0.371 | 0.114 |
| BGA | Bangladeshi Garole | 24 | 0.286 | 0.307 | 0.069 |
| BGE | Bangladeshi East BGE | 24 | 0.270 | 0.313 | 0.140 |
| BHM | Black Headed Mountain | 24 | 0.330 | 0.340 | 0.029 |
| BMN | Morada Nova | 22 | 0.310 | 0.319 | 0.028 |
| BOS | Bundner Oberlander Sheep | 24 | 0.359 | 0.347 | −0.034 |
| BRL | Border Leicester | 48 | 0.290 | 0.291 | 0.001 |
| BSI | SantaInes | 47 | 0.345 | 0.353 | 0.023 |
| CAS | Castellana | 23 | 0.378 | 0.375 | −0.008 |
| CFT | Cyprus Fat Tail | 30 | 0.326 | 0.315 | −0.036 |
| CHA | Changthangi | 29 | 0.332 | 0.353 | 0.060 |
| CHI | Chios | 23 | 0.324 | 0.330 | 0.019 |
| CHU | Churra | 120 | 0.359 | 0.362 | 0.008 |
| CME | Chinese Merino | 23 | 0.365 | 0.359 | −0.017 |
| COM | Comisana | 24 | 0.372 | 0.368 | −0.009 |
| CPW | Australian Coopworth | 19 | 0.378 | 0.366 | −0.033 |
| DJA | Djallonke | 11 | 0.293 | 0.300 | 0.024 |
| DSH | Dorset Horn | 21 | 0.315 | 0.296 | −0.065 |
| EFB | East Friesian Brown | 39 | 0.295 | 0.300 | 0.018 |
| EFW | East Friesian White | 9 | 0.316 | 0.310 | −0.018 |
| EMZ | Ethiopian Menz | 34 | 0.321 | 0.325 | 0.013 |
| ERS | Engadine Red Sheep | 24 | 0.369 | 0.366 | −0.009 |
| FIN | Finnsheep | 99 | 0.345 | 0.356 | 0.031 |
| GAL | Galway | 49 | 0.335 | 0.334 | −0.003 |
| GAR | Indian Garole | 26 | 0.288 | 0.295 | 0.024 |
| GCN | Gulf Coast Native | 94 | 0.364 | 0.377 | 0.034 |
| GTX | German Texel | 46 | 0.349 | 0.354 | 0.016 |
| GUR | Garut | 22 | 0.333 | 0.332 | −0.003 |
| IDC | Deccani | 24 | 0.335 | 0.337 | 0.005 |
| ISF | Irish Suffolk | 55 | 0.315 | 0.330 | 0.046 |
| KRS | Karakas | 18 | 0.355 | 0.347 | −0.022 |
| LAD | Ladoum | 12 | 0.313 | 0.319 | 0.019 |
| LEC | Leccese | 24 | 0.346 | 0.372 | 0.070 |
| LME | Lacaune (meat) | 78 | 0.365 | 0.371 | 0.016 |
| LMI | Lacaune (milk) | 103 | 0.361 | 0.365 | 0.009 |
| MCM | Macarthur Merino | 10 | 0.231 | 0.229 | −0.011 |
| MLA | Merinolandschaf | 24 | 0.362 | 0.359 | −0.007 |
| MOG | Moghani | 34 | 0.363 | 0.363 | −0.001 |
| NDZ | Norduz | 20 | 0.352 | 0.337 | −0.046 |
| NQA | Namaqua Afrikaner | 12 | 0.287 | 0.252 | −0.140 |
| NTX | NewZealand Texel | 24 | 0.344 | 0.347 | 0.008 |
| OJA | Ojalada | 24 | 0.376 | 0.377 | 0.001 |
| ONS | Old Norwegianspaelsau | 15 | 0.339 | 0.358 | 0.054 |
| PEU | Peul | 12 | 0.333 | 0.345 | 0.035 |
| QEZ | Qezel | 35 | 0.361 | 0.366 | 0.015 |
| RAA | Rasaaragonesa | 22 | 0.384 | 0.383 | −0.002 |
| RDA | Ronderib Afrikaner | 17 | 0.315 | 0.300 | −0.048 |
| RMA | Red Maasai | 45 | 0.324 | 0.323 | −0.004 |
| RMB | Merinos de Rambouillet | 102 | 0.345 | 0.357 | 0.032 |
| ROM | New Zealand Romney | 24 | 0.347 | 0.355 | 0.023 |
| SAB | SardDSHinian Ancestral Black | 20 | 0.355 | 0.339 | −0.049 |
| SBF | Scottish Blackface | 56 | 0.361 | 0.360 | −0.002 |
| SBS | Swiss Black | 24 | 0.354 | 0.352 | −0.006 |
| SKZ | Sakiz | 22 | 0.333 | 0.313 | −0.065 |
| SMS | Swiss Mirror | 24 | 0.356 | 0.350 | −0.015 |
| SPC | Spael | 3 | 0.334 | 0.335 | 0.001 |
| SPW | Spael | 32 | 0.330 | 0.334 | 0.012 |
| STE | St Elizabeth | 10 | 0.373 | 0.371 | −0.005 |
| STX | ScottishTexel | 80 | 0.349 | 0.331 | −0.053 |
| SUM | Sumatra | 24 | 0.303 | 0.315 | 0.037 |
| SWA | Swiss White Alpine | 24 | 0.354 | 0.351 | −0.008 |
| TIB | Tibetan | 37 | 0.323 | 0.341 | 0.052 |
| TOU | Touabire | 12 | 0.339 | 0.347 | 0.025 |
| VBS | Valais Blacknose | 24 | 0.303 | 0.318 | 0.049 |
| VRS | Valais Red | 24 | 0.323 | 0.314 | −0.031 |
| WIL | Wiltshire | 23 | 0.269 | 0.267 | −0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Missohou, A.; Kaboré, B.; Flori, L.; Ayssiwede, S.B.; Hornick, J.-L.; Raes, M.; Cabaraux, J.-F. Analysis of the Genetic Diversity and Population Structure of Four Senegalese Sheep Breeds Using Medium-Density Single-Nucleotide Polymorphisms. Animals 2022, 12, 1512. https://doi.org/10.3390/ani12121512
Missohou A, Kaboré B, Flori L, Ayssiwede SB, Hornick J-L, Raes M, Cabaraux J-F. Analysis of the Genetic Diversity and Population Structure of Four Senegalese Sheep Breeds Using Medium-Density Single-Nucleotide Polymorphisms. Animals. 2022; 12(12):1512. https://doi.org/10.3390/ani12121512
Chicago/Turabian StyleMissohou, Ayao, Basse Kaboré, Laurence Flori, Simplice Bosco Ayssiwede, Jean-Luc Hornick, Marianne Raes, and Jean-François Cabaraux. 2022. "Analysis of the Genetic Diversity and Population Structure of Four Senegalese Sheep Breeds Using Medium-Density Single-Nucleotide Polymorphisms" Animals 12, no. 12: 1512. https://doi.org/10.3390/ani12121512
APA StyleMissohou, A., Kaboré, B., Flori, L., Ayssiwede, S. B., Hornick, J.-L., Raes, M., & Cabaraux, J.-F. (2022). Analysis of the Genetic Diversity and Population Structure of Four Senegalese Sheep Breeds Using Medium-Density Single-Nucleotide Polymorphisms. Animals, 12(12), 1512. https://doi.org/10.3390/ani12121512






