Differential Gene Expression Associated with Soybean Oil Level in the Diet of Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Diets
2.3. Fatty Acid Profile
2.4. Tissue RNA Extraction and RNA Sequencing
2.5. Data Analysis, Differentially Expressed Genes, and Functional Enrichment Analysis
3. Results
3.1. Fatty Acid Profile for Skeletal Muscle and Liver Tissue
3.2. Sequencing Data and Differential Expression Analysis
3.3. Common Differentially Expressed Genes between Skeletal Muscle and Liver Tissue
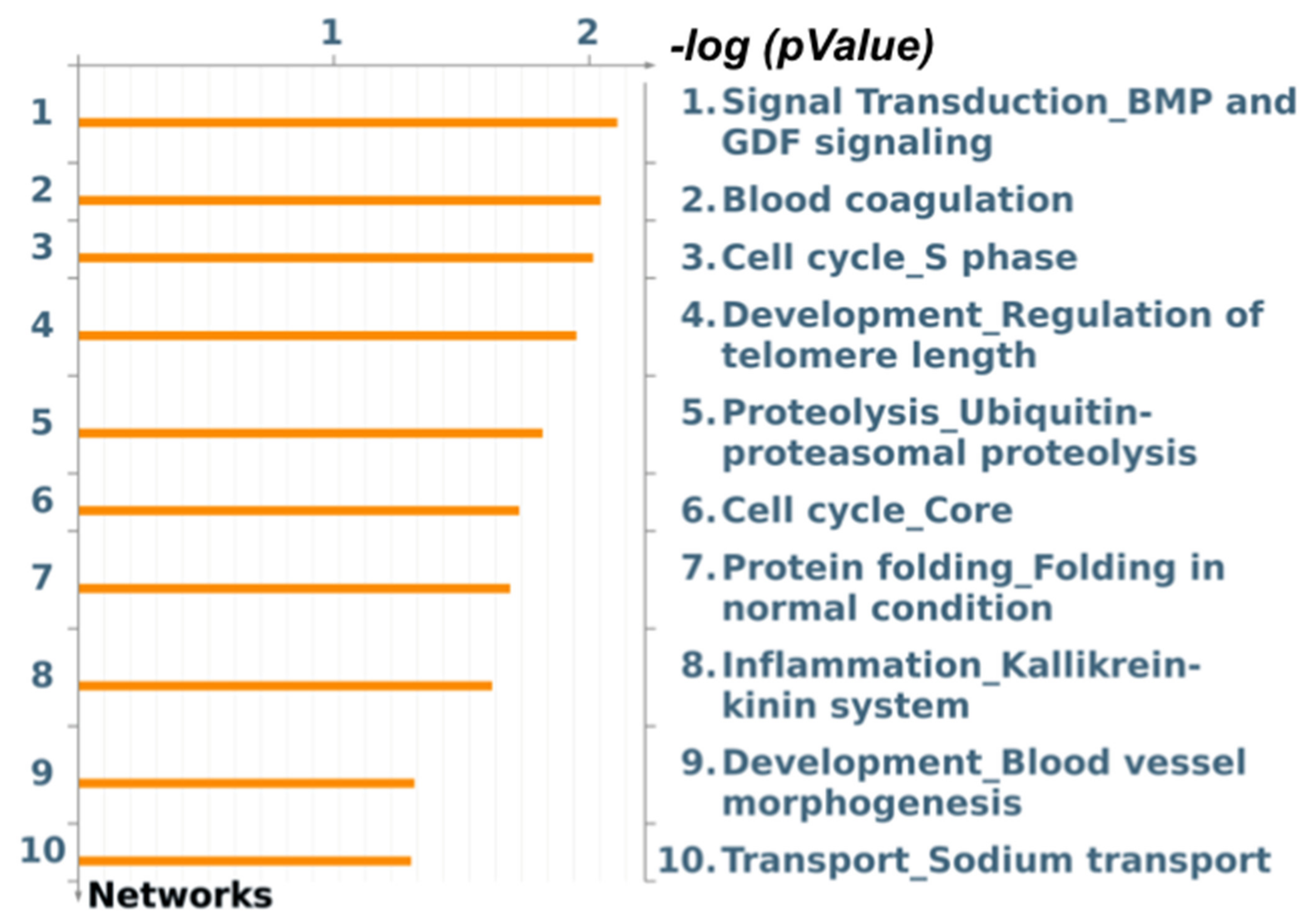
3.4. Functional Enrichment Analysis for Skeletal Muscle Differential Expression
3.5. Functional Enrichment Analysis for Liver Differential Expression
4. Discussion
4.1. Different Levels of Dietary Soybean Oil Modulates Fat Deposition
4.2. Different Levels of Dietary Soybean Oil Modulate Gene Expression in Skeletal Muscle
4.3. Soybean Oil Added to Pig’s Diet Modulates Gene Expression in Liver Tissue
4.4. Genes Common to Dietary Treatments and Overview of the Effect of Soybean Oil Addition in Different Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization Europe WHO/Europe. Noncommunicable Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 20 November 2021).
- Litten-Brown, J.C.; Corson, A.M.; Clarke, L. Porcine models for the metabolic syndrome, digestive and bone disorders: A general overview. Animals 2010, 4, 899–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Xing, M.; Gu, X. Research Progress on Oxidative Stress and Its Nutritional Regulation Strategies in Pigs. Animals 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Yao, Y.; Yin, H.; Cai, Z.; Wang, Y.; Bai, L.; Kern, C.; Halstead, M.; Chanthavixay, G.; Trakooljul, N.; et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Malgwi, I.H.; Halas, V.; Grünvald, P.; Schiavon, S.; Jócsák, I. Genes Related to Fat Metabolism in Pigs and Intramuscular Fat Content of Pork: A Focus on Nutrigenetics and Nutrigenomics. Animals 2022, 12, 150. [Google Scholar] [CrossRef]
- Li, R.; Jia, Z.; Trush, M.A. Defining ROS in biology and medicine. React. Oxyg. Species 2016, 1, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Laaksonen, D.E.; Nyyssönen, K.; Niskanen, L.; Rissanen, T.H.; Salonen, J.T. Prediction of Cardiovascular Mortality in Middle-aged Men by Dietary and Serum Linoleic and Polyunsaturated Fatty Acids. Arch. Intern. Med. 2005, 165, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Lunney, J.K. Advances in Swine Biomedical Model Genomics. Int. J. Biol. Sci. 2007, 3, 179–184. [Google Scholar] [CrossRef]
- Kragh, P.M.; Nielsen, A.L.; Li, J.; Du, Y.; Lin, L.; Schmidt, M.; Bøgh, I.B.; Holm, I.E.; Jakobsen, J.E.; Johansen, M.G.; et al. Hemizygous minipigs produced by random gene insertion and handmade cloning express the Alzheimer’s disease-causing dominant mutation APPsw. Transgenic Res. 2009, 18, 545–558. [Google Scholar] [CrossRef]
- Reddy, A.M.; Zheng, Y.; Jagadeeswaran, G.; Macmil, S.L.; Graham, W.B.; A Roe, B.; Desilva, U.; Zhang, W.; Sunkar, R. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genom. 2009, 10, 65. [Google Scholar] [CrossRef] [Green Version]
- Ramayo-Caldas, Y.; Mach, N.; Esteve-Codina, A.; Corominas, J.; Castelló, A.; Ballester, M.; Estellé, J.; Ibáñez-Escriche, N.; I Fernández, A.; Pérez-Enciso, M.; et al. Liver transcriptome profile in pigs with extreme phenotypes of intramuscular fatty acid composition. BMC Genom. 2012, 13, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, Y.; Paton, C.M. Lipid metabolic features of skeletal muscle in pathological and physiological conditions. In Lipid Signaling and Metabolism; Academic Press: Cambridge, MA, USA, 2020; pp. 359–383. [Google Scholar] [CrossRef]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bülow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardi, S.; Marcuzzi, A.; Piscianz, E.; Tommasini, A.; Fabris, B. The Complex Interplay between Lipids, Immune System and Interleukins in Cardio-Metabolic Diseases. Int. J. Mol. Sci. 2018, 19, 4058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotamisligil, G.S. Foundations of Immunometabolism and Implications for Metabolic Health and Disease. Immunity 2017, 47, 406–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.; Nara, T.; Nakamura, M.; Nara, T. Essential fatty acid synthesis and its regulation in mammals. Prostaglandins Leukot. Essent. Fat. Acids 2003, 68, 145–150. [Google Scholar] [CrossRef]
- Wood, J.D.; Enser, M.; Fisher, A.V.; Nute, G.R.; Sheard, P.R.; Richardson, R.I.; Whittington, F.M.; Hughes, S.I. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Hoffmann, G. Association of dietary, circulating, and supplement fatty acids with coronary risk. Ann. Intern. Med. 2014, 160, 398–406. [Google Scholar] [CrossRef]
- Cesar, A.S.M.; Regitano, L.C.A.; Poleti, M.D.; Andrade, S.C.S.; Tizioto, P.C.; Oliveira, P.S.N.; Felício, A.M.; do Nascimento, M.L.; Chaves, A.S.; Lanna, D.P.D.; et al. Differences in the skeletal muscle transcriptome profile associated with extreme values of fatty acids content. BMC Genom. 2016, 17, 961. [Google Scholar] [CrossRef] [Green Version]
- Schmid, A. The Role of Meat Fat in the Human Diet. Crit. Rev. Food Sci. Nutr. 2010, 51, 50–66. [Google Scholar] [CrossRef]
- Kritchevsky, D. Antimutagenic and some other effects of conjugated linoleic acid. Br. J. Nutr. 2000, 83, 459–465. [Google Scholar] [CrossRef]
- Terés, S.; Barceló-Coblijn, G.; Benet, M.; Álvarez, R.; Bressani, R.; Halver, J.E.; Escribá, P.V. Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. USA 2008, 105, 13811–13816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, E.K. The Protective Effect of the Mediterranean Diet: Focus on Cancer and Cardiovascular Risk. Med. Princ. Pract. 2011, 20, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Tsimikas, S.; Reaven, P.D. The role of dietary fatty acids in lipoprotein oxidation and atherosclerosis. Curr. Opin. Lipidol. 1998, 9, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Park, J.C.; Kim, S.C.; Lee, S.H.; Jang, H.C.; Kim, N.K.; Jung, H.J.; Kim, I.C.; Seong, H.H.; Choi, B.H. Effects of Dietary Fat Types on Growth Performance, Pork Quality, and Gene Expression in Growing-finishing Pigs. Asian-Australas. J. Anim. Sci. 2012, 25, 1759–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alencar, S.A.D.S.; Kiefer, C.; Nascimento, K.M.R.D.S.; Viana, L.H.; Corassa, A.; Gomes, M.D.N.B.; Marçal, D.A.; Farias, T.V.A. Dietary soybean oil modulates fatty acid composition of pork. Trop. Anim. Health Prod. 2021, 53, 1–7. [Google Scholar] [CrossRef]
- Federation of Animal Science Societies. Fass Guide for the Care and Use of Agricultural Animals in Research and Teaching, 3rd ed.; Federation of Animal Science Societies: Champaign, IL, USA, 2010. [Google Scholar]
- Fujii, J.; Otsu, K.; Zorzato, F.; de Leon, S.; Khanna, V.K.; Weiler, J.E.; O’Brien, P.J.; MacLennan, D.H. Identification of a Mutation in Porcine Ryanodine Receptor Associated with Malignant Hyperthermia. Science 1991, 253, 448–451. [Google Scholar] [CrossRef]
- Almeida, V.; Silva, J.; Schinckel, A.; Meira, A.; Moreira, G.; Gomes, J.; Poleti, M.; Dargelio, M.; Patinho, I.; Contreras-Castillo, C.; et al. Effects of increasing dietary oil inclusion from different sources on growth performance, carcass and meat quality traits, and fatty acid profile in genetically lean immunocastrated male pigs. Livest. Sci. 2021, 248, 104515. [Google Scholar] [CrossRef]
- Fanalli, S.L.; da Silva, B.P.M.; Gomes, J.D.; de Almeida, V.V.; Afonso, J.; Reecy, J.M.; Koltes, J.E.; Koltes, D.; de Carvalho Baileiro, J.C.; Freitas, L.; et al. Effect of Dietary Soybean Oil Inclusion on Liver-Related Transcription Factors in a Pig Model for Metabolic Diseases. Sci. Rep. 2022, 12, 10318. [Google Scholar] [CrossRef]
- Rostagno, H.S. Tabelas brasileiras para aves e suínos: Composição de alimentos e exigências nutricionais. Tabelas Bras. para aves e suínos Compos. Aliment. Exig. Nutr. 2011, 2, 186. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- AOCS. Rapid Determination of Oil/Fat Utilizing High Temperature Solvent Extraction; Official approved procedure Am 5-04; AOCS Press: Urbana, IL, USA, 2005. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- MetaCore. MetaCore and Cortellis solution [Internet] (Clarivate Analytics, London, UK) 2022. Available online: https://portal.genego.com/ (accessed on 20 November 2021).
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Hoy, A.J. Lipid metabolism in skeletal muscle: Generation of adaptive and maladaptive intracellular signals for cellular function. Am. J. Physiol. Metab. 2012, 302, E1315–E1328. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Maruyama, T.; Yoshikawa, N.; Matsumiya, R.; Ma, Y.; Ito, N.; Tasaka, Y.; Kuribara-Souta, A.; Miyata, K.; Oike, Y.; et al. A muscle-liver-fat signalling axis is essential for central control of adaptive adipose remodelling. Nat. Commun. 2015, 6, 6693. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, P.; Leray, V.; Diez, M.; Serisier, S.; Le Bloc’H, J.; Siliart, B.; Dumon, H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008, 92, 272–283. [Google Scholar] [CrossRef]
- Vitali, M.; Sirri, R.; Zappaterra, M.; Zambonelli, P.; Giannini, G.; Fiego, D.P.L.; Davoli, R. Functional analysis finds differences on the muscle transcriptome of pigs fed an n-3 PUFA-enriched diet with or without antioxidant supplementations. PLoS ONE 2019, 14, e0212449. [Google Scholar] [CrossRef]
- Benítez, R.; Trakooljul, N.; Núñez, Y.; Isabel, B.; Murani, E.; De Mercado, E.; Gómez-Izquierdo, E.; García-Casco, J.; López-Bote, C.; Wimmers, K.; et al. Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources. Genes 2019, 10, 589. [Google Scholar] [CrossRef] [Green Version]
- Adeola, O.; Bajjalieh, N.L. Energy concentration of high-oil corn varieties for pigs. J. Anim. Sci. 1997, 75, 430–436. [Google Scholar] [CrossRef]
- De la Llata, M.; Dritz, S.S.; Tokach, M.D.; Goodband, R.D.; Nelssen, J.L.; Loughin, T.M. Economics of increasing lysine: Calorie ratio and dietary fat addition in growing-finishing pigs reared in a commercial environment. J. Swine Health 2001, 9, 215–223. [Google Scholar]
- Zhang, L.; Wang, X.; Chen, S.; Wang, S.; Tu, Z.; Zhang, G.; Zhu, H.; Li, X.; Xiong, J.; Liu, Y. Medium-Chain Triglycerides Attenuate Liver Injury in Lipopolysaccharide-Challenged Pigs by Inhibiting Necroptotic and Inflammatory Signaling Pathways. Int. J. Mol. Sci. 2018, 19, 3697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Zhou, Y.; Jiang, S.; Huang, F.; Peng, J.; Jiang, S. Transcriptional response of porcine skeletal muscle to feeding a linseed-enriched diet to growing pigs. J. Anim. Sci. Biotechnol. 2016, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enser, M.; Richardson, R.; Wood, J.; Gill, B.; Sheard, P. Feeding linseed to increase the n-3 PUFA of pork: Fatty acid composition of muscle, adipose tissue, liver and sausages. Meat Sci. 2000, 55, 201–212. [Google Scholar] [CrossRef]
- Kelson, T.L.; McVoy, J.R.S.; Rizzo, W.B. Human liver fatty aldehyde dehydrogenase: Microsomal localization, purification, and biochemical characterization. Biochim. Biophys. Acta (BBA)—Gen. Subj. 1997, 1335, 99–110. [Google Scholar] [CrossRef]
- Peters-Golden, M.; Gleason, M.M.; Togias, A. Cysteinyl leukotrienes: Multi-functional mediators in allergic rhinitis. Clin. Exp. Allergy 2006, 36, 689–703. [Google Scholar] [CrossRef] [Green Version]
- Maslov, L.N.; Naryzhnaya, N.V.; Boshchenko, A.A.; Popov, S.V.; Ivanov, V.V.; Oeltgen, P.R. Is oxidative stress of adipocytes a cause or a consequence of the metabolic syndrome? J. Clin. Transl. Endocrinol. 2019, 15, 1–5. [Google Scholar] [CrossRef]
- Pan, C.; Xing, J.-H.; Zhang, C.; Zhang, Y.-M.; Zhang, L.-T.; Wei, S.-J.; Zhang, M.-X.; Wang, X.-P.; Yuan, Q.-H.; Xue, L.; et al. Aldehyde dehydrogenase 2 inhibits inflammatory response and regulates atherosclerotic plaque. Oncotarget 2016, 7, 35562–35576. [Google Scholar] [CrossRef] [Green Version]
- Bazewicz, C.G.; Dinavahi, S.S.; Schell, T.D.; Robertson, G.P. Aldehyde dehydrogenase in regulatory T-cell development, immunity and cancer. Immunology 2018, 156, 47–55. [Google Scholar] [CrossRef]
- Chu, A.; Najafzadeh, P.; Sullivan, P.; Cone, B.; Elshimali, R.; Shakeri, H.; Janzen, C.; Mah, V.; Wadehra, M. Aldehyde dehydrogenase isoforms and inflammatory cell populations are differentially expressed in term human placentas affected by intrauterine growth restriction. Placenta 2019, 81, 9–17. [Google Scholar] [CrossRef]
- Krahmer, N.; Farese, R.V., Jr.; Walther, T.C. Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Gohda, T.; Makita, Y.; Shike, T.; Tanimoto, M.; Funabiki, K.; Horikoshi, S.; Tomino, Y. Identification of Epistatic Interaction Involved in Obesity Using the KK/Ta Mouse as a Type 2 Diabetes Model: Is Zn-α2 glycoprotein-1 a candidate gene for obesity? Diabetes 2003, 52, 2175–2181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzanavari, T.; Bing, C.; Trayhurn, P. Postnatal expression of zinc-α2-glycoprotein in rat white and brown adipose tissue. Mol. Cell. Endocrinol. 2007, 279, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Jager, J.; Grémeaux, T.; Gonzalez, T.; Bonnafous, S.; Debard, C.; Laville, M.; Vidal, H.; Tran, A.; Gual, P.; Le Marchand-Brustel, Y.; et al. Tpl2 Kinase Is Upregulated in Adipose Tissue in Obesity and May Mediate Interleukin-1β and Tumor Necrosis Factor-α Effects on Extracellular Signal–Regulated Kinase Activation and Lipolysis. Diabetes 2010, 59, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faulconnier, Y.; Bernard, L.; Boby, C.; Domagalski, J.; Chilliard, Y.; Leroux, C. Extruded linseed alone or in combination with fish oil modifies mammary gene expression profiles in lactating goats. Animals 2018, 12, 1564–1575. [Google Scholar] [CrossRef]
- Ollier, S.; Robert-Granié, C.; Bernard, L.; Chilliard, Y.; Leroux, C. Mammary Transcriptome Analysis of Food-Deprived Lactating Goats Highlights Genes Involved in Milk Secretion and Programmed Cell Death. J. Nutr. 2007, 137, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Hirai, K.; Hussey, H.J.; Barber, M.D.; A Price, S.; Tisdale, M.J. Biological evaluation of a lipid-mobilizing factor isolated from the urine of cancer patients. Cancer Res. 1998, 58, 2359–2365. [Google Scholar]
- Choi, J.-W.; Liu, H.; Mukherjee, R.; Yun, J.W. Downregulation of Fetuin-B and Zinc-a2-glycoprotein is Linked to Impaired Fatty Acid Metabolism in Liver Cells. Cell. Physiol. Biochem. 2012, 30, 295–306. [Google Scholar] [CrossRef]
- Liu, T.; Luo, X.; Li, Z.-H.; Wu, J.-C.; Luo, S.-Z.; Xu, M.-Y. Zinc-α2-glycoprotein 1 attenuates non-alcoholic fatty liver disease by negatively regulating tumour necrosis factor-α. World J. Gastroenterol. 2019, 25, 5451–5468. [Google Scholar] [CrossRef]
- Morris, D.L.; Cho, K.W.; DelProposto, J.L.; Oatmen, K.E.; Geletka, L.M.; Martinez-Santibanez, G.; Singer, K.; Lumeng, C.N. Adipose Tissue Macrophages Function As Antigen-Presenting Cells and Regulate Adipose Tissue CD4+ T Cells in Mice. Diabetes 2013, 62, 2762–2772. [Google Scholar] [CrossRef] [Green Version]
- Feraco, A.; Gorini, S.; Armani, A.; Camajani, E.; Rizzo, M.; Caprio, M. Exploring the Role of Skeletal Muscle in Insulin Resistance: Lessons from Cultured Cells to Animal Models. Int. J. Mol. Sci. 2021, 22, 9327. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.-G.; Ko, H.J.; Cho, Y.-R.; Kim, H.-J.; Ma, Z.; Yu, T.Y.; Friedline, R.H.; Kurt-Jones, E.; Finberg, R.; Fischer, M.A.; et al. Interleukin-10 Prevents Diet-Induced Insulin Resistance by Attenuating Macrophage and Cytokine Response in Skeletal Muscle. Diabetes 2009, 58, 2525–2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varma, V.; Yao-Borengasser, A.; Rasouli, N.; Nolen, G.T.; Phanavanh, B.; Starks, T.; Gurley, C.; Simpson, P.; McGehee, R.E., Jr.; Kern, P.A.; et al. Muscle inflammatory response and insulin resistance: Synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1300–E1310. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Fatty acids and inflammation: The cutting edge between food and pharma. Eur. J. Pharmacol. 2011, 668 (Suppl. S1), S50–S58. [Google Scholar] [CrossRef] [PubMed]
- Djuricic, I.; Calder, P. Beneficial Outcomes of Omega-6 and Omega-3 Polyunsaturated Fatty Acids on Human Health: An Update for 2021. Nutrients 2021, 13, 2421. [Google Scholar] [CrossRef] [PubMed]
- Innes, J.K.; Calder, P.C. Omega-6 fatty acids and inflammation. Prostaglandins Leukot. Essent. Fat. Acids 2018, 132, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Horodyska, J.; Hamill, R.M.; Reyer, H.; Trakooljul, N.; Lawlor, P.G.; McCormack, U.M.; Wimmers, K. RNA-Seq of Liver from Pigs Divergent in Feed Efficiency Highlights Shifts in Macronutrient Metabolism, Hepatic Growth and Immune Response. Front. Genet. 2019, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Carroll, J.B.; Bates, G.; Steffan, J.; Saft, C.; Tabrizi, S.J. Treating the whole body in Huntington’s disease. Lancet Neurol. 2015, 14, 1135–1142. [Google Scholar] [CrossRef]
- Felipo, V. Hepatic encephalopathy: Effects of liver failure on brain function. Nat. Rev. Neurosci. 2013, 14, 851–858. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Chern, Y.; Juo, C.-G. The dysfunction of hepatic transcriptional factors in mice with Huntington’s Disease. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2011, 1812, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Sroka, K.; Voigt, A.; Deeg, S.; Reed, J.C.; Schulz, J.B.; Bähr, M.; Kermer, P. BAG1 modulates huntingtin toxicity, aggregation, degradation, and subcellular distribution. J. Neurochem. 2009, 111, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.K.; McLean, P.J. Molecular Chaperones as Rational Drug Targets for Parkinsons Disease Therapeutics. CNS Neurol. Disord.—Drug Targets 2010, 9, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Turturici, G.; Sconzo, G.; Geraci, F. Hsp70 and Its Molecular Role in Nervous System Diseases. Biochem. Res. Int. 2011, 2011, 618127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nollen, E.A.A.; Brunsting, J.F.; Song, J.; Kampinga, H.; Morimoto, R.I. Bag1 Functions In Vivo as a Negative Regulator of Hsp70 Chaperone Activity. Mol. Cell. Biol. 2000, 20, 1083–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adrie, C.; Monchi, M.; Fulgencio, J.-P.; Cottias, P.; Haouache, H.; Alvarez-Gonzalvez, A.; Guerrini, P.; Cavaillon, J.-M.; Adib-Conquy, M. Immune Status and apoptosis activation during brain death. Shock 2010, 33, 353–362. [Google Scholar] [CrossRef]
- Jana, N.; Zemskov, E.A.; Wang, G.-H.; Nukina, N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 2001, 10, 1049–1059. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.-Z.; Zhang, J.-W.; Zheng, S. What we know about ST13, a co-factor of heat shock protein, or a tumor suppressor? J. Zhejiang Univ. Sci. B 2007, 8, 170–176. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Song, L.; Zhu, P.; Zhang, B.; Tao, Y.; Xu, X.; Li, F.; Wu, K.; Liang, J.; Shao, D.; et al. Single-Cell Exome Sequencing and Monoclonal Evolution of a JAK2-Negative Myeloproliferative Neoplasm. Cell 2012, 148, 873–885. [Google Scholar] [CrossRef] [Green Version]
- De Freitas, R.C.C.; Bortolin, R.H.; Lopes, M.B.; Hirata, M.H.; Hirata, R.D.C.; Silbiger, V.N.; Luchessi, A.D. Integrated analysis of miRNA and mRNA gene expression microarrays: Influence on platelet reactivity, clopidogrel response and drug-induced toxicity. Gene 2016, 593, 172–178. [Google Scholar] [CrossRef]
- Finnerty, J.R.; Wang, W.X.; Hébert, S.S.; Wilfred, B.R.; Mao, G.; Nelson, P.T. The miR-15/107 group of MicroRNA genes: Evolutionary biology, cellular functions, and roles in human diseases. J. Mol. Biol. 2010, 402, 491–509. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.-X. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur. J. Neurosci. 2005, 22, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Signore, S.C.; Wouters, F.S.; Schmitz, M.; Baehr, M.; Kermer, P. BAG1 Overexpression Stabilizes High Molecular Tau Protein—A Crucial Role of the Co-chaperone in Tau Pathology. Am. J. Psychiatry Neurosci. 2021, 9, 77. [Google Scholar] [CrossRef]
- Elliott, E.; Laufer, O.; Ginzburg, I. BAG-1M is up-regulated in hippocampus of Alzheimer’s disease patients and associates with tau and APP proteins. J. Neurochem. 2009, 109, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Dong, S.; Gu, F.; Hu, Y.; Zhao, Z. Advances in the Pathogenesis of Alzheimer’s Disease: Focusing on Tau-Mediated Neurodegeneration. Transl. Neurodegener. 2012, 1, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustelo, X.R. Regulatory and Signaling Properties of the Vav Family. Mol. Cell. Biol. 2000, 20, 1461–1477. [Google Scholar] [CrossRef] [Green Version]
- Baer, A.S.; Syed, Y.A.; Kang, S.U.; Mitteregger, D.; Vig, R.; Ffrench-Constant, C.; Franklin, R.J.M.; Altmann, F.; Lubec, G.; Kotter, M.R. Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain 2009, 132, 465–481. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, M.; Ishizaki, T.; Boku, S.; Watanabe, N.; Fujita, A.; Iwamatsu, A.; Obinata, T.; Ohashi, K.; Mizuno, K.; Narumiya, S. Signaling from Rho to the Actin Cytoskeleton Through Protein Kinases ROCK and LIM-kinase. Science 1999, 285, 895–898. [Google Scholar] [CrossRef]
- Rego, A.C.; Oliveira, C.R. Mitochondrial Dysfunction and Reactive Oxygen Species in Excitotoxicity and Apoptosis: Implications for the Pathogenesis of Neurodegenerative Diseases. Neurochem. Res. 2003, 28, 1563–1574. [Google Scholar] [CrossRef] [Green Version]
- Beal, M. Mitochondrial dysfunction in neurodegenerative diseases. Biochim. Biophys. Acta 1998, 1366, 211–223. [Google Scholar] [CrossRef] [Green Version]
- Brustovetsky, N.; Brustovetsky, T.; Purl, K.J.; Capano, M.; Crompton, M.; Dubinsky, J.M. Increased Susceptibility of Striatal Mitochondria to Calcium-Induced Permeability Transition. J. Neurosci. 2003, 23, 4858–4867. [Google Scholar] [CrossRef] [Green Version]
- Steffan, J.S.; Kazantsev, A.; Spasic-Boskovic, O.; Greenwald, M.; Zhu, Y.-Z.; Gohler, H.; Wanker, E.E.; Bates, G.P.; Housman, D.E.; Thompson, L.M. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 2000, 97, 6763–6768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Sekiya, M.; Najima, Y.; Okazaki, S.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Inoue, N.; et al. p53 Involvement in the Pathogenesis of Fatty Liver Disease. J. Biol. Chem. 2004, 279, 20571–20575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, V. Bradykinin. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: New York, NY, USA, 2007; pp. 1–3. [Google Scholar]
| Fatty Acid (%) | Dietary Treatment | |||
|---|---|---|---|---|
| SOY1.5 | SOY3.0 | Pooled SEM 1 | p-Value | |
| Saturated fatty acid (SFA) | ||||
| Myristic acid (C14:0) | 1.14 | 1.19 | 0.04 | 0.20 |
| Palmitic acid (C16:0) | 25.50 | 25.01 | 0.21 | 0.21 |
| Stearic acid (C18:0) | 12.18 | 11.89 | 0.15 | 0.42 |
| Monounsaturated fatty acid (MUFA) | ||||
| Palmitoleic acid (C16:1) | 2.86 | 3.17 | 0.13 | 0.02 |
| Eicosenoic acid (C20:1) | 0.51 | 0.55 | 0.03 | 0.11 |
| Oleic acid (C18:1 n-9) | 38.93 | 44.15 | 1.40 | <0.01 |
| Polyunsaturated fatty acid (PUFA) | ||||
| Linoleic acid (C18:2 n-6) | 17.90 | 13.28 | 1.12 | <0.01 |
| Alpha-linolenic acid (C18:3 n-3) | 0.77 | 0.56 | 0.06 | <0.01 |
| Eicosapentaenoic acid (C20:5 n-3) | 0.30 | 0.15 | 0.09 | 0.12 |
| Docosahexaenoic acid (C22:6 n-3) | 0.36 | 0.16 | 0.08 | 0.03 |
| Total SFA | 38.83 | 38.09 | 0.65 | 0.26 |
| Total MUFA | 42.29 | 47.70 | 1.48 | <0.01 |
| Total PUFA | 19.28 | 14.80 | 1.72 | 0.02 |
| Total n-3 PUFA 2 | 1.35 | 0.87 | 0.15 | <0.01 |
| Total n-6 PUFA 3 | 17.90 | 13.28 | 1.12 | <0.01 |
| PUFA:SFA ratio 4 | 0.50 | 0.39 | 0.05 | 0.03 |
| n-6:n-3 PUFA ratio 5 | 14.20 | 17.29 | 1.70 | 0.10 |
| Atherogenic index | 0.49 | 0.48 | 0.09 | 0.43 |
| Fatty Acid (%) | Dietary Treatment | Pooled SEM 1 | p-Value | |
|---|---|---|---|---|
| SOY1.5 | SOY3.0 | |||
| Saturated fatty acid (SFA) | ||||
| Myristic acid (C14:0) | 0.73 | 0.98 | 0.05 | <0.01 |
| Palmitic acid (C16:0) | 20.92 | 22.98 | 0.40 | <0.01 |
| Stearic acid (C18:0) | 25.48 | 21.28 | 1.06 | <0.01 |
| Monounsaturated fatty acid (MUFA) | ||||
| Palmitoleic acid (C16:1) | 0.66 | 0.93 | 0.05 | <0.01 |
| Oleic acid (C18:1 n-9) | 21.36 | 27.84 | 1.06 | <0.01 |
| Polyunsaturated fatty acid (PUFA) | ||||
| Linoleic acid (C18:2 n-6) | 27.02 | 23.64 | 0.67 | <0.01 |
| Alpha-linolenic acid (C18:3 n-3) | 1.42 | 1.17 | 0.10 | 0.07 |
| Eicosapentaenoic acid (C20:5 n-3, EPA) | 0.58 | 0.27 | 0.11 | 0.04 |
| Docosahexaenoic acid (C22:6 n-3, DHA) | 1.18 | 0.99 | 0.11 | 0.17 |
| Total SFA | 46.69 | 45.24 | 1.03 | 0.31 |
| Total MUFA | 22.01 | 28.78 | 1.04 | <0.01 |
| Total PUFA | 30.79 | 26.06 | 0.55 | <0.01 |
| Total n-3 PUFA 2 | 3.75 | 2.42 | 0.37 | <0.01 |
| Total n-6 PUFA 3 | 27.02 | 23.64 | 0.67 | <0.01 |
| PUFA:SFA ratio 4 | 0.67 | 0.58 | 0.02 | <0.01 |
| n-6:n-3 PUFA ratio 5 | 8.51 | 9.90 | 0.50 | 0.05 |
| Atherogenic index | 0.42 | 0.51 | 0.01 | <0.01 |
| Gene Common | Description | Reference |
|---|---|---|
| ENSSSCG00000009578 Cyclin-dependent kinase 20 (CDK20) | Cell-cycle-related kinase. Its expression is related to the activation of β-catenin-TCF signaling and cell cycle progression. Can activate cyclin-dependent kinase 2 which is related to cell growth. | [38,39] |
| ENSSSCG00000014903 Coiled-coil domain-containing 90B (CCDC90B) | Paralog of the MCUR1 gene (Mitochondrial Calcium Uniporter Regulator 1) which is related to the Ca, cAMP, and lipid-signaling pathways. | [39] |
| ENSSSCG00000022842 LOC100525692 | Protein-encoding gene. | [39] |
| ENSSSCG00000022842 Alpha-1,3-Glucosyltransferase (ALG6) | Related to N-linked glycosylation. | [39] |
| ENSSSCG00000017914 Glycolipid Transfer Protein Domain-Containing Protein 2 GLTPD2 | Participates in the transfer of glycolipids. | [39] |
| ENSSSCG00000051557 | - | - |
| Pathway Maps | p-Value | DEG ¹ |
|---|---|---|
| Fatty acid omega oxidation | 0.0333 | AL3A2 |
| Leukotriene 4 biosynthesis and metabolism | 0.0442 | AL3A2 |
| TNF-alpha, IL-1 beta induces dyslipidemia and inflammation in obesity and type 2 diabetes in adipocytes | 0.0464 | AZGP1 |
| Breakdown of CD4+ T cell peripheral tolerance in type 1 diabetes mellitus | 0.0539 | CD4 |
| Triacylglycerol metabolism p.1 | 0.0656 | AL3A2 |
| Oxidative stress in adipocyte dysfunction in type 2 diabetes and metabolic syndrome X | 0.0699 | AL3A2 |
| Peroxisomal branched-chain fatty acid oxidation | 0.0908 | AL3A2 |
| Process Networks | p-Value | DEG ¹ |
|---|---|---|
| Chemostaxis | 0.0018 | CCR10, GPCRs, CD4 |
| Cell adhesion_Leucocyte chemostaxis | 0.00378 | CCR10, GPCRs, CD4 |
| Immune response_Antigen presentation | 0.0046 | CD4, AZGP1 |
| Signal transduction_Leptin signaling | 0.0156 | A2M, T-A2MG |
| Inflammation_Kallikrein–kinin system | 0.0443 | A2M, T-A2MG |
| Reproduction_Male sex differentiation | 0.0699 | Tektin 1, AKAP3 |
| Pathway Maps | p-Value | DEG ¹ |
|---|---|---|
| HSP70 and HSP40-dependent folding in Huntington’s disease | 0.01034 | BAG-1, ST13 (Hip) |
| Inhibition of remyelination in multiple sclerosis: regulation of cytoskeleton proteins | 0.03022 | MAPT, MELC |
| Tau pathology in Alzheimer disease | 0.04543 | MAPT, PP2C |
| Mitochondrial dysfunction in neurodegenerative diseases | 0.05153 | ANT |
| Dual role of p53 in transcription deregulation in Huntington’s Disease | 0.07179 | p21 |
| LRRK2 in neuronal apoptosis in Parkinson’s disease | 0.09869 | ANT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanalli, S.L.; da Silva, B.P.M.; Gomes, J.D.; de Almeida, V.V.; Freitas, F.A.O.; Moreira, G.C.M.; Silva-Vignato, B.; Afonso, J.; Reecy, J.; Koltes, J.; et al. Differential Gene Expression Associated with Soybean Oil Level in the Diet of Pigs. Animals 2022, 12, 1632. https://doi.org/10.3390/ani12131632
Fanalli SL, da Silva BPM, Gomes JD, de Almeida VV, Freitas FAO, Moreira GCM, Silva-Vignato B, Afonso J, Reecy J, Koltes J, et al. Differential Gene Expression Associated with Soybean Oil Level in the Diet of Pigs. Animals. 2022; 12(13):1632. https://doi.org/10.3390/ani12131632
Chicago/Turabian StyleFanalli, Simara Larissa, Bruna Pereira Martins da Silva, Julia Dezen Gomes, Vivian Vezzoni de Almeida, Felipe André Oliveira Freitas, Gabriel Costa Monteiro Moreira, Bárbara Silva-Vignato, Juliana Afonso, James Reecy, James Koltes, and et al. 2022. "Differential Gene Expression Associated with Soybean Oil Level in the Diet of Pigs" Animals 12, no. 13: 1632. https://doi.org/10.3390/ani12131632






