First Complete Cytochrome B Sequences and Molecular Taxonomy of Bat Species from Sri Lanka
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
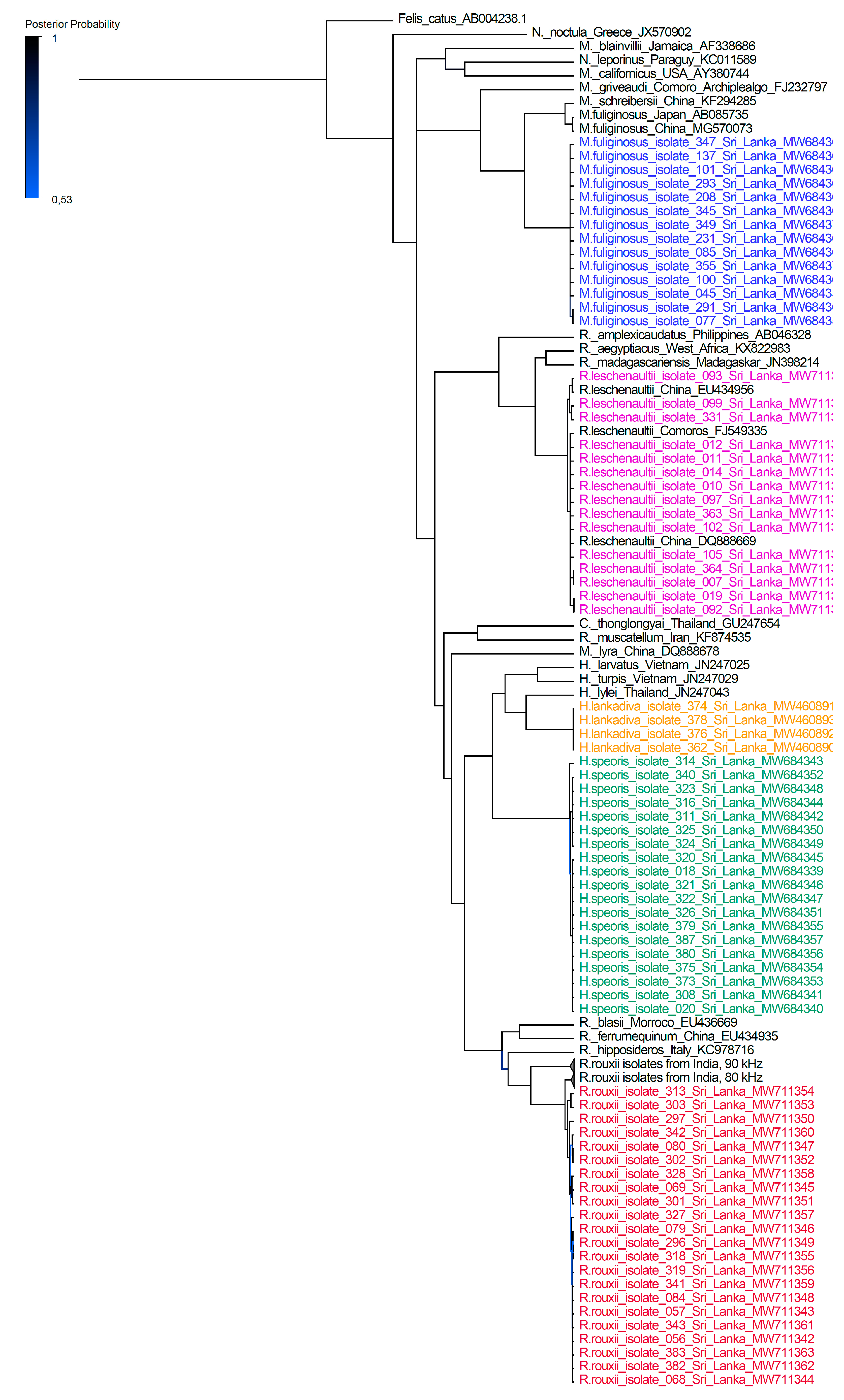
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Higher Taxonomy. Available online: https://www.mammaldiversity.org/taxa.html (accessed on 26 June 2022).
- Yapa, W. A Field Guide to the Bats of Sri Lanka; Dilmah Ceylon Tea Company PLC: Colombo, Sri Lanka, 2017. [Google Scholar]
- Koopman, K.F. Mammal Species of the World, a Taxonomic and Geographic Reference; Wilson, D.E., Reeder, D.M., Eds.; Smithsonian Institution Press: Washington, DC, USA, 1993; pp. 137–241. [Google Scholar]
- Jones, G.; Teeling, E. The Evolution of Echolocation in Bats. Trends Ecol. Evol. 2006, 21, 149–156. [Google Scholar] [CrossRef]
- Wei, K.; Zhang, T.; Ma, L. Divergent and Convergent Evolution of Housekeeping Genes in Human-Pig Lineage. PeerJ 2018, 6, e4840. [Google Scholar] [CrossRef]
- Mapatuna, Y.; Gunasekera, M.; Rathnasooriya, W.; Goonesekere, N.; Bates, P. Unravelling the Taxonomic Status of the Genus Cynopterus (Chiroptera: Pteropodidae) in Sri Lanka by Multivariate Morphometrics and Mitochondrial DNA Sequence Analysis. Mamm. Biol. 2002, 67, 321–337. [Google Scholar] [CrossRef]
- Yapa, W. Ecological and Ethological Studies on Cave Dwelling Bats in Sri Lanka; University of Colombo: Colombo, Sri Lanka, 1992. [Google Scholar]
- Sikes, R. 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Chattopadhyay, B.; Garg, K.M.; Vinoth Kumar, A.K.; Paramanantha Swami Doss, D.; Ramakrishnan, U.; Kandula, S. Sibling Species in South Indian Populations of the Rufous Horse-Shoe Bat Rhinolophus Rouxii. Conserv. Genet. 2012, 13, 1435–1445. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Kelaart, E.F. Prodromus Faunae Zeylanicae: Being Contributions to the Zoology of Ceylon. Kandy; The Observer Press: Colombo, Sri Lanka, 1852. [Google Scholar]
- Phillips, W.W.A. Manual of the Mammals of Ceylon; Dulau and Company: London, UK, 1935. [Google Scholar]
- Weerakoon, D.; Wijesundara, S. The National Red List 2012 of Sri Lanka; Biodiversity Secretariat of the Ministry of Environment and Natural Herbarium, Dept. of National Botanic Gardens: Peradeniya, Sri Lanka, 2012. [Google Scholar]
- Yapa, W.B.; Digana, P.M.C.B.; Ratnasooriya, W.D.; Rübsamen, R.; Costa, H.H.; Randeniya, P.V. Breeding Associated Migration of Miniopterus Schreibersii between Two Natural Caves in Sri Lanka. In Proceedings of the 19th Annual Sessions of the Institute of Biology, Colombo, Sri Lanka; Institute of Biology: Colombo, Sri Lanka, 1998; p. 21. [Google Scholar]
- Chattopadhyay, B.; Schuller, G.; Garg, K.M.; Kandula, S. A New Phonic Type of the Rufous Horseshoe Bat Rhinolophus Rouxii from Southern India. Curr. Sci. 2010, 99, 114–118. [Google Scholar]
- Neuweiler, G.; Metzner, W.; Heilmann, U.; Rübsamen, R.; Eckrich, M.; Costa, H.H. Foraging Behaviour and Echolocation in the Rufous Horseshoe Bat (Rhinolophus rouxi) of Sri Lanka. Behav. Ecol. Sociobiol. 1987, 20, 53–67. [Google Scholar] [CrossRef]
- Hassanin, A.; Bonillo, C.; Tshikung, D.; Pongombo Shongo, C.; Pourrut, X.; Kadjo, B.; Nakouné, E.; Tan Tu, V.; Prié, V.; Goodman, S.M. Phylogeny of African Fruit Bats (Chiroptera, Pteropodidae) Based on Complete Mitochondrial Genomes. J. Zool. Syst. Evol. Res. 2020, 58, 1395–1410. [Google Scholar] [CrossRef]
- Luis, A.D.; Hayman, D.T.S.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.C.; Mills, J.N.; Timonin, M.E.; Willis, C.K.R.; Cunningham, A.A.; et al. A Comparison of Bats and Rodents as Reservoirs of Zoonotic Viruses: Are Bats Special? Proc. Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef] [Green Version]
- Wibbelt, G.; Moore, M.S.; Schountz, T.; Voigt, C.C. Emerging Diseases in Chiroptera: Why Bats? Biol. Lett. 2010, 6, 438–440. [Google Scholar] [CrossRef] [Green Version]
- Perera, H.I.T.; Yapa, W.B.; Perera, H.K.K. Isolation of Salmonella Species in Rousettus Leschenaultii Fruit Bats in Sri Lanka; Institute of Biology: Colombo, Sri Lanka, September 2016; p. 33. [Google Scholar]
- Hu, B.; Ge, X.; Wang, L.F.; Shi, Z. Bat Origin of Human Coronaviruses. Virol. J. 2015, 12, 221. [Google Scholar] [CrossRef] [Green Version]
- Muzeniek, T.; Perera, T.; Siriwardana, S.; Bas, D.; Kaplan, F.; Öruc, M.; Becker-Ziaja, B.; Schwarz, F.; Premawansa, G.; Premawansa, S.; et al. Detection of Alpha- and Betacoronaviruses in Miniopterus fuliginosus and Rousettus leschenaultii, Two Species of Sri Lankan Bats. Vaccines 2021, 9, 650. [Google Scholar] [CrossRef]
- Muzeniek, T.; Perera, T.; Siriwardana, S.; Bas, D.; Kaplan, F.; Öruc, M.; Becker-Ziaja, B.; Perera, I.; Weerasena, J.; Handunnetti, S.; et al. Full Genome of BatCoV/MinFul/2018/SriLanka, a Novel Alpha-Coronavirus Detected in Miniopterus Fuliginosus, Sri Lanka. Viruses 2022, 14, 337. [Google Scholar] [CrossRef]
- Muzeniek, T.; Perera, T.; Siriwardana, S.; Bayram, F.; Bas, D.; Öruc, M.; Becker-Ziaja, B.; Perera, I.; Weerasena, J.; Handunnetti, S.; et al. Paramyxovirus Diversity within One Population of Miniopterus Fuliginosus Bats in Sri Lanka. Pathogens 2022, 11, 434. [Google Scholar] [CrossRef]
- Calisher, C. Bats: Important Hosts of Emerging Viruses. Int. J. Antimicrob. Agents 2007, 29, S80–S81. [Google Scholar] [CrossRef]
- Bonilla-Aldana, D.K.; Jimenez-Diaz, S.D.; Arango-Duque, J.S.; Aguirre-Florez, M.; Balbin-Ramon, G.J.; Paniz-Mondolfi, A.; Suárez, J.A.; Pachar, M.R.; Perez-Garcia, L.; Delgado-Noguera, L.A.; et al. Bats in Ecosystems and Their Wide Spectrum of Viral Infectious Potential Threats: SARS-CoV-2 and Other Emerging Viruses. Int. J. Infect. Dis. 2021, 102, 87–96. [Google Scholar] [CrossRef]
- Longdon, B.; Brockhurst, M.A.; Russell, C.A.; Welch, J.J.; Jiggins, F.M. The Evolution and Genetics of Virus Host Shifts. PLoS Pathog. 2014, 10, e1004395. [Google Scholar] [CrossRef] [Green Version]
- Arnaout, Y.; Djelouadji, Z.; Robardet, E.; Cappelle, J.; Cliquet, F.; Touzalin, F.; Jimenez, G.; Hurstel, S.; Borel, C.; Picard-Meyer, E. Genetic Identification of Bat Species for Pathogen Surveillance across France. PLoS ONE 2022, 17, e0261344. [Google Scholar] [CrossRef]
- De Benedictis, P.; Leopardi, S.; Markotter, W.; Velasco-Villa, A. The Importance of Accurate Host Species Identification in the Framework of Rabies Surveillance, Control and Elimination. Viruses 2022, 14, 492. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perera, T.; Schwarz, F.; Muzeniek, T.; Siriwardana, S.; Becker-Ziaja, B.; Perera, I.C.; Handunnetti, S.; Weerasena, J.; Premawansa, G.; Premawansa, S.; et al. First Complete Cytochrome B Sequences and Molecular Taxonomy of Bat Species from Sri Lanka. Animals 2022, 12, 1674. https://doi.org/10.3390/ani12131674
Perera T, Schwarz F, Muzeniek T, Siriwardana S, Becker-Ziaja B, Perera IC, Handunnetti S, Weerasena J, Premawansa G, Premawansa S, et al. First Complete Cytochrome B Sequences and Molecular Taxonomy of Bat Species from Sri Lanka. Animals. 2022; 12(13):1674. https://doi.org/10.3390/ani12131674
Chicago/Turabian StylePerera, Thejanee, Franziska Schwarz, Therese Muzeniek, Sahan Siriwardana, Beate Becker-Ziaja, Inoka C. Perera, Shiroma Handunnetti, Jagathpriya Weerasena, Gayani Premawansa, Sunil Premawansa, and et al. 2022. "First Complete Cytochrome B Sequences and Molecular Taxonomy of Bat Species from Sri Lanka" Animals 12, no. 13: 1674. https://doi.org/10.3390/ani12131674
APA StylePerera, T., Schwarz, F., Muzeniek, T., Siriwardana, S., Becker-Ziaja, B., Perera, I. C., Handunnetti, S., Weerasena, J., Premawansa, G., Premawansa, S., Nitsche, A., Yapa, W., & Kohl, C. (2022). First Complete Cytochrome B Sequences and Molecular Taxonomy of Bat Species from Sri Lanka. Animals, 12(13), 1674. https://doi.org/10.3390/ani12131674






