1. Introduction
Early gut colonization and succession of microorganisms play a significant role in animal health, the feed conversion rate, and feed intake, and is of great significance for animals to grow and develop during their whole life. In human and mouse model studies, the host is dependent on the distal gut microbiome to provide the capacity for carbohydrate structure digestion and absorption, the modulation of bile acid conjugative patterns, fat emulsification and absorption, and the modulation of lipid metabolism. However, the nutrient and energy value of food is not absolute, but is affected partly by the digestive capacity of an individual’s microbiota [
1,
2,
3]. Nevertheless, early studies of the rumen microbial communities focused on the types and function of microorganisms that affect the host’s production performance, blood physiological parameters, and rumen development, using both culture and genetic approaches; however, we found that relatively few studies have been published regarding intestinal tract microorganism [
4]. Therefore, studying the relationship between gut microbes with nutrient digestibility is critical to improve production performance, aiming to reduce farming costs and increase benefits to lambs.
The digestive system of ruminants is inhabited by many species and types of microorganisms, and their main role is decomposition of nutrients [
5]. In the early part of life, both rumen and intestinal microbes undergo rapid changes in colonization, succession, and stabilization. The dominance of strictly anaerobic species very soon after birth (2 d) and the early appearance (first week) of populations of cellulolytic and methanogenic bacteria, show that the rumen ecosystem is quickly established after birth before the rumen itself becomes functional [
6]. The microbial community and the function of rumen are not well-established before 20 days of age; therefore, there is a degree of plasticity in the rumen bacterial community during the first 20 days of post-natal development in lambs, and this might provide an opportunity for interventions to improve rumen fermentation [
7]. Significantly, Furman et al. observed that members of the core successional microbiome appeared earlier than all others, within the first 140 days of life, with most of them being introduced during the first days after birth in cattle [
8]. The intestinal microbiota is a complex community of microorganisms that live in the intestinal tract, playing an important role in barrier function and providing many benefits for the host, including defending against pathogens, using all available nutrients, and secreting compounds that kill or inhibit unwelcome organisms that would compete for nutrients [
9]. Studies have shown that homeostasis of gastrointestinal micro-organisms is threatened by many external factors, the most important of which are heat stress, psychological stress, environment, and diet [
10]. Furthermore, different weaning strategies have different effects on the gastrointestinal microflora of early lambs [
11].
The development of the nutrient digestibility of lambs is very important for their whole life. The early postnatal and weaning stages are key for lamb digestive tract development [
12]. However, the interaction and causal links between the gastrointestinal microflora and the digestibility of the host are still uncertain. On the one hand, the intake of all nutrients increases with age, and with the increase of solid feed intake, intestinal microbial diversity index and bacterial abundance change significantly before 49 days of age [
13]. On the other hand, gastrointestinal microbes play an important role in the digestion and absorption of nutrients. Research has shown that a higher abundance of functional bacteria in the rumen improves forage digestibility, while producing high concentrations of NH
3-N and short volatile fatty acids (VFAs) to rapidly improve growth performance [
14]. For example, Prevotella bacteria appear to be associated with propionic acid production and play a pivotal role in degrading and utilizing plant non-cellulosic polysaccharides, protein, starch, and xylans [
15]. Bacteria such as Verrucomicrobia contain a wide range of glycoside hydrolases, which play an important role in the decomposition of polysaccharides and cellobiose [
16]. Researchers have been mainly focused on understanding the rumen microbiota’s contribution to the host in the last decade. However, each region of the gastrointestinal is spatially specialized depending on factors including physiology substrate availabilities, retention time of digesta, and pH levels [
17]. These factors are all expected to have a profound impact on the local microbial assemblages and functions, thereby affecting the digestive, immunological, metabolic, and endocrinological processes in ruminants [
18]. The rectal fecal flora of ruminants is highly similar to that of the colon and cecum [
19], and fecal samples are easy to collect. Increased understanding of the relationship between fecal microorganisms and nutrient utilization in ruminants has a positive effect on reducing feed cost and improving animal productivity. However, it remains to be further explored whether the colonization of the intestinal flora of young lambs has an important effect on the development of nutrient digestion and absorption.
Thus, the objective of this study was to evaluate the periodical changes of growth, nutrient intake, digestibility, fecal nutrient content, and fecal microbiota, and to explore the relationship between microbial diversity and nutrient digestibility in early lambs. We hypothesized that with the increase of starter intake, nutrient digestibility changes rapidly, which alters the composition of fermentation substrates and thus affects the fecal microbiota.
2. Materials and Methods
2.1. Experiment Design and Animal Management
According to the birth weight principle (mean ± SD: 3.29 ± 0.68 kg), six healthy neonatal male Hu lambs were selected from a commercial sheep farm (Jinchang Zhongtian Sheep Industry Co. Ltd., Jinchang, China). From birth to 3 days, the lambs were kept indoors with ewes to ensure adequate colostrum intake. The lambs were trained to use the nipple bottle containing reconstructed milk replacer (crude protein 23.22%, fat 13.20%, air-dried basis; Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China;
Table 1) from 4 to 6 days. Milk replacer was reconstituted at 200 g/L in water and provided for lambs at a temperature of 40 °C. Each lamb was fed 50 mL of milk replacer three times a day (at 09:00, 15:00, and 21:00). At 7 days old, the lambs were separated from ewes and placed in individual pens (0.65 m × 1.10 m), and artificial feeding with milk replacer (2% of average body weight per day) completely replaced lactation, and the starter diet was added. At 21 days old, the lambs were fed with complete starter and milk replacer was stopped. These diets, published in China, are formulated to meet the requirements of the feeding standard of meat-producing sheep and goats (NYT816-2004), and their formula and nutritional composition are shown in
Table 1. All lambs had free access to the starter diet and clean water. All lambs were weighed every 14 days to calculate average body weight and average daily gain. The starter intake of each lamb was recorded daily as the difference between offered and refused feed and the average intake was calculated.
2.2. Sample Collection and Measurement of Nutrient Digestion
Fecal samples for total microbial DNA extraction were obtained by rectal stimulation before morning feeding from lambs at 7, 21, 35, and 49 days old, and were stored in sterile test tubes at −80 °C. Apparent total tract digestibility was determined by total fecal collection method [
20]. At 7–10, 18–21, 32–35, and 46–49 days old, the starter intake of each lamb was recorded daily as the difference between offered and refused feed, and all feces were collected and separated from urine with a slatted floor and gauze. The feces were weighed daily and, pooled for each 3-d period. A proportion of the feces was divided, and stored in 10% sulfuric acid for crude protein (CP) determination. Other fecal samples were dried at 65 °C to calculate the initial moisture and stored at room temperature for dry matter (DM), crude fat ether extract (EE), neutral detergent fiber (NDF), acid detergent fiber (ADF), and starch determination. The milk replacer, starter and feces were analyzed for DM(drying at 105 °C), CP(AOAC International, 2000), EE(AOAC International, 2000) [
21], NDF and ADF following a previously described method with heat-stable alpha-amylase and sodium sulfate used in the NDF procedure [
22], and starch using a commercial assay kit (Solarbio, Shanghai, China) according to the manufacturer’s instructions. The apparent digestibility of protein, starch, fat, DM, ADF, and NDF was calculated in the following Equation:
where AD is the apparent digestibility of protein, starch, fat, DM, ADF, or NDF (%), Fi is the intake of protein, starch, fat, DM, ADF or NDF (g), and Ff is the fecal output of protein, starch, fat, DM, ADF or NDF (g).
2.3. Bacterial DNA Extraction
DNA from different samples was extracted by using an E.Z.N.A. ®Stool DNA Kit (D4015, Omega Bio-tek, Norcross, GA, USA) according to manufacturer’s instructions. The kit contains a reagent designed to recover DNA from trace amounts of sample and has been shown to be effective for the preparation of DNA of most bacteria. Nuclear-free water was used as the blank control. The total DNA was eluted in 50 µL of Elution buffer and stored at −80 °C until measurement by using PCR by LC-Bio Technology Co., Ltd., HangZhou, China.
2.4. PCR Amplification and 16S rDNA Sequencing
The V4 region of the prokaryotic (bacterial and archaeal) small-subunit (16S) rRNA gene was amplified with primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The 5′ ends of the primers were tagged with specific barcodes per sample and with universal sequencing primers.
PCR amplification was performed in a total volume of 25 µL. The reaction mixture contained 25 ng of template DNA, 12.5 µL of PCR Premix, 2.5 µL of each primer, and PCR-grade water to adjust the volume. The PCR conditions to amplify the prokaryotic 16S fragments consisted of an initial denaturation at 98 °C for 30 s; 35 cycles of denaturation at 98 °C for 10 s, annealing at 54 °C/52 °C for 30 s, and extension at 72 °C for 45 s; and a final extension at 72 °C for 10 min. The PCR products were confirmed by using 2% agarose gel electrophoresis. Throughout the DNA extraction process, ultrapure water, instead of a sample solution, was used to exclude the possibility of false-positive PCR results, as a negative control. The PCR products were purified by using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by using a Qubit instrument (Invitrogen, Waltham, MA, USA). Amplicon pools were prepared for sequencing and the size and quantity of the amplicon library were assessed on an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and with a Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, USA), respectively. A PhiX Control library (v3) (Illumina Inc., San Diego, CA, USA) was combined with the amplicon library. The libraries were sequenced either as 250-bp paired-end reads (250PE) MiSeq runs, and one library was sequenced with both protocols by using the standard Illumina sequencing primers, eliminating the need for a third (or fourth) index read. Samples were sequenced on the Illumina MiSeq platform according to the manufacturer’s recommendations, provided by LC-Bio. The sequencing data were deposited into the Sequence Read Archive (SRA) of NCBI and can be accessed via accession number PRJNA836702.
2.5. Sequence and Statistical Analysis
Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged by using FLASH (32.0.0.371). Quality filtering on the raw tags were performed under specific filtering conditions to obtain high-quality clean tags, according to FastQC (V 0.10.1). Chimeric sequences were filtered by using Verseach software (v2.3.4). Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs) by using Verseach (v2.3.4). Representative sequences were chosen for each OTU, and taxonomic data were then assigned to each representative sequence by using the RDP (Ribosomal Database Project) classifier. The differences in the dominant species in the different groups were identified and multiple sequence alignment was conducted by using the PyNAST software to study the phylogenetic relationship of the different OTUs. OTU abundance information was normalized by using the number of sequences in the sample with the least number of sequences as a standard. Alpha diversity analysis was applied to assess the complexity of species diversity for a sample by using four indices: Chao1, Shannon, Simpson, and Observed species. These indices were calculated by using QIIME (Version 1.8.0). Beta diversity analysis was used to evaluate differences in species complexity in the samples. Beta diversity was calculated by principal coordinates analysis (PCoA) and cluster analysis by QIIME software (Version 1.8.0).
The data of microbial diversity indices (among these diversity indices, the Shannon index measures uncertainty about the identity of species in the sample, and its units quantify information, while the Simpson measures a probability, specifically, the probability that two individuals, drawn randomly from the sample, will be of different species [
23]. Coverage is the proportion of individuals belonging to undiscovered species in a community that can be reliably estimated based solely on the frequency of species already present in the sample [
24]. Chao1 was asymptotic richness estimators and could predict the community diversity [
25]), bacterial densities, growth performance, apparent digestibility, daily intake, apparent digestion, daily excretion, and fecal nutrient contents were analyzed by using one-way ANOVA and the least significant difference (LSD) post hoc tests in SPSS software (version 25.0; IBM Corp., Armonk, NY, USA) with 5 degrees of freedom. The following statistical model was used:
Yij = μ + Ai + eij, where
Y is the microbial diversity indices, bacterial densities, growth performance, apparent digestibility, daily intake, apparent digestion, daily excretion or fecal nutrient contents;
μ is the mean;
A is the age; and
e is the residual error. Spearman correlation coefficients were used to evaluate relationships among the most abundant genera and nutrient contents in feces and nutrient digestion using the R software (version 4.1.1). Statistical significance was set at
p < 0.05,
p < 0.001 indicated an extremely significant difference.
4. Discussion
Fecal microbes can represent the intestinal flora to study the relationship between the periodic changes of intestinal flora and digestibility in early lambs. Previous research has shown that that fecal microbiota is highly variable in the early life of calves [
26], playing an important role in body health [
27], digestive processes, and immune response, and is affected by many factors, such as age [
28], diet [
28], the environment [
29], and weaning [
11]. Furthermore, a study in house mice observed that 93.3% of OTUs were shared between fecal and lower gastrointestinal samples [
30]. Another study indicated that gastrointestinal origin is a primary determinant for the fecal microbiota composition [
31]. These results indicated that fecal samples have good potential to identify microbial members derived from the digestive tract. Therefore, a more detailed understanding of the progression of the early lamb fecal microbiome from the neonatal period, including weaning and the commencement of starter feed, will provide insights into what constitutes a stable microbiome at these crucial stages of growth and development.
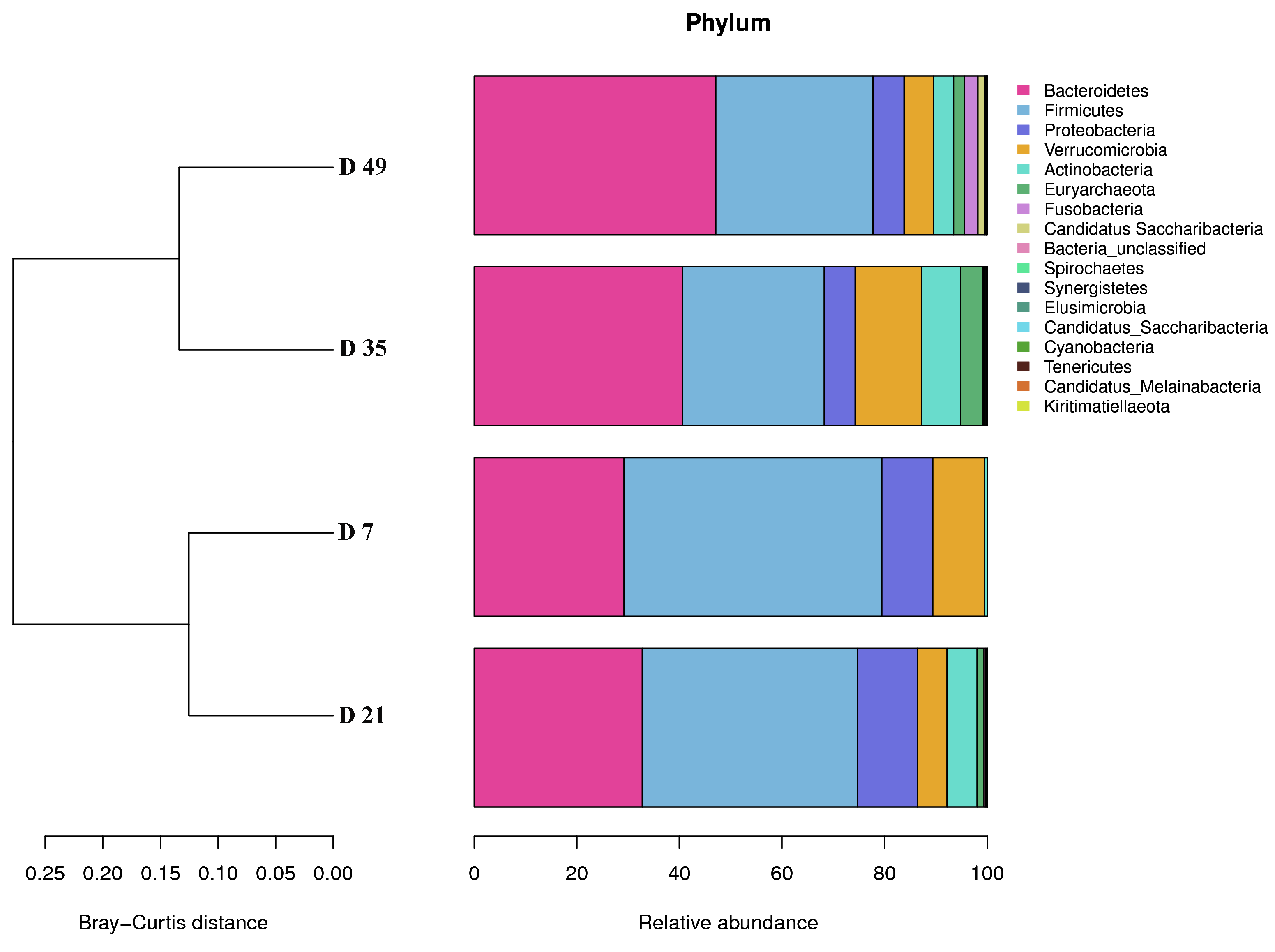
The diversity of fecal flora showed an increasing trend until the end of the experiment. In this study, alpha diversity indices (Observed species, Shannon, Simpson and Chao1) of the fecal microbiota increased with age until the end of the experiment, which corresponded with the process of microbial colonization and development observed in the gut of ruminants [
32]. A highly diverse gut microbiota is generally considered beneficial for host health and is regarded as a sign of a mature gut microbiota [
33]. However, we observed a decreased alpha diversity during the early lambs after weaning (D21), the main reason for which might be sudden diet transition from milk replacer to solid feed after weaning [
34]. Therefore, the development of the intestinal microflora of lambs up to 49 days old is a key stage, in which lambs drink less milk (highly digestible) and more starter (less digestible) over time, and microbes updated to change in nutrients available in the intestinal tract.
Transformation to solid feed changes the fecal microflora. Consistent with previous studies, this study demonstrated that Bacteroidetes and Firmicutes were the two most dominant phyla in the fecal microbial communities of early lambs. A study based on human infants indicated that Bacteroidetes and Firmicutes were the most prevalent phyla [
35]. Our results revealed decreases in the relative abundances of five phyla (Firmicutes, Verrucomicrobia, Proteobacteria, and Synergistetes) and increases in the relative abundances of four phyla (Bacteroidetes, Actinobacteria, Euryarchaeota and Fusobacteria) with the age of early lambs. These microorganisms changed mainly because of introduction of fiber in the solid feed. This result might be related to the gradual maturation and stabilization of the intestinal flora. At the genus level, the relative abundances of 8 genera declined, whereas the relative abundances of 12 microbe genera increased with increasing age and weaning of the early lambs. The relative abundances of only four genera changed dramatically:
Lachnospiraceae and
Clostridium XlVa numbers decreased, whereas
Akkermansia and
Clostridium XlVb numbers increased significantly. Reports suggested that the presence of
Clostridium XlVa might be the main cause of diarrhea in early lambs [
36,
37]. Furthermore, proteolysis is common among
Clostridia species [
38]; therefore, the changes in the levels of these species might have been be caused by the decrease in the protein composition in milk after weaning and increased body resistance with the growth of lambs. In addition,
Lachnospiraceae ferment diverse plant polysaccharides to short-chain fatty acids and alcohols [
39], thus the decline in abundance of
Lachnospiraceae_unclassified might be related to the gradual increase in fibrous feed intake. Indeed, the results showed that the abundance of Lachnospiraceae_unclassified correlated negatively with the fecal fiber content. Interestingly, the abundance of some bacteria decreased suddenly after weaning and then gradually increased toward D49, such as
lactobacillus and
Bacteroidetes. By contrast,
Akkermansia suddenly increased after weaning and the gradually decreased toward D49. These results suggested that these genera are susceptible to the changes in diet composition.
Bifidobacterium and
Lactobacillus are representative probiotic bacteria. Both genera have been proven to beneficially affect intestinal health through different mechanisms and have anti-proliferative, proapoptotic, and anti-oxidant properties [
40]. After a period of time, their abundance gradually recovered, indicating that the microflora adapted to change in diet composition after weaning.
With the increase in feed intake, the intake and daily digestion of DM, protein, starch, NDF, and ADF increased gradually; however, their digestibility decreased gradually. The apparent digestibility of DM, CP, starch, fat, NDF, and ADF significantly decreased by 21.77%, 23.15%, 23.62%, 19.38%, and 45.24%, respectively, from 7 to 35 days of age. This is consistent with the change rule of nutrient digestibility [
12], which proposes that increased fiber results in a reduction of apparent nutrient digestibility [
41,
42]. Furthermore, we analyzed the correlation between nutrient digestibility and the fecal microflora, and the correlation between fecal nutrient contents and the fecal microflora, respectively, to explore the interaction and causality between host nutrient digestion and the microflora. Starch showed no statistical significance and correlation. Studies have reported that the main culprit is within the rumen, where more than 90% [
43] of dietary plant cell walls and 20~90% of the starch are degraded [
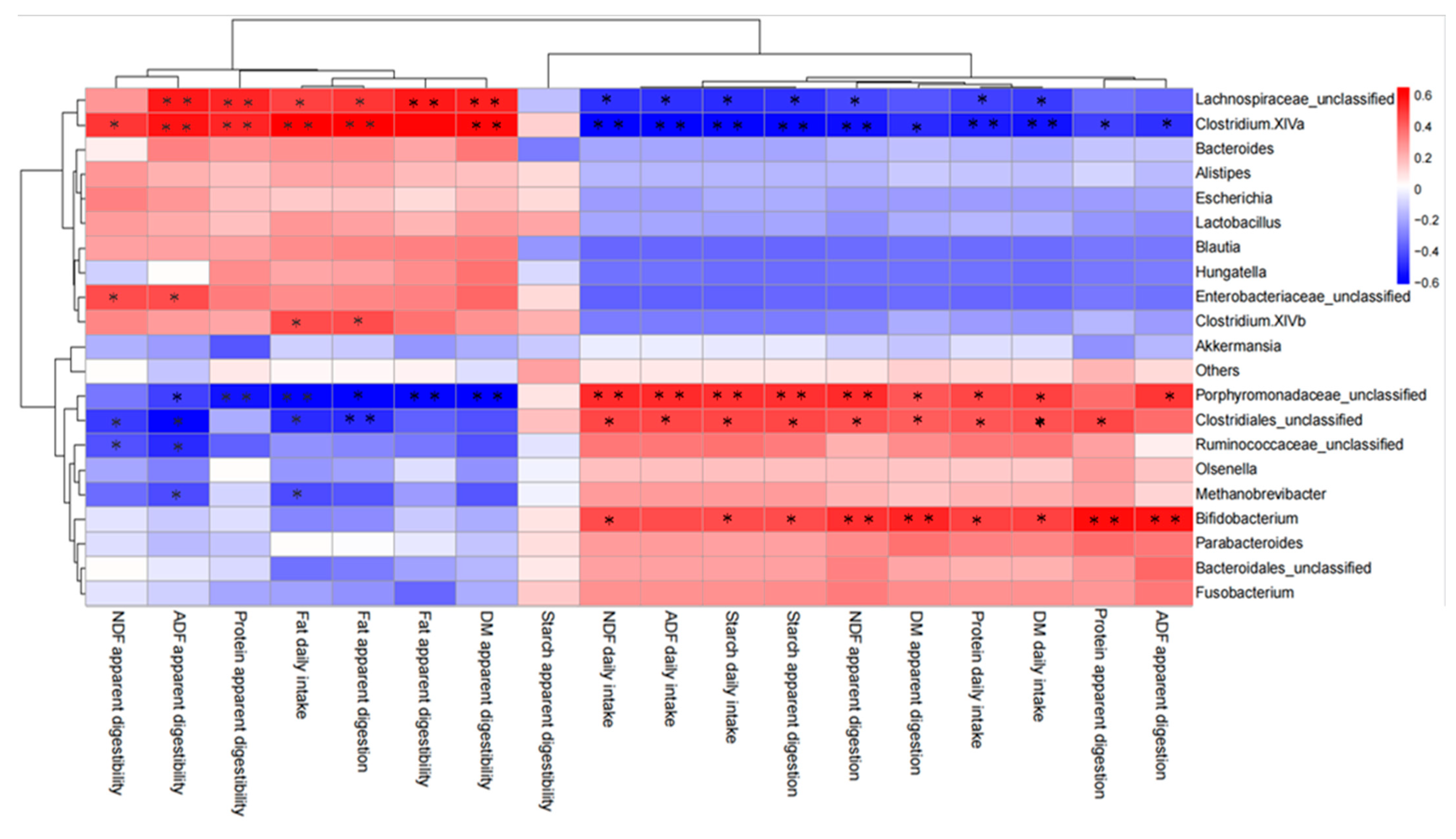
44], whereas nutrients entering the gut comprised recalcitrant carbohydrates. The abundances of
Parabacteroides,
Olsenella,
Methanobrevibacter,
Porphyromonadaceae_unclassified, and
Clostridiales_unclassified were correlated significantly and negatively with fecal DM, protein, and fat contents, but positively with NDF and ADF contents, suggesting that the fecal fiber content affected the abundance of these bacteria, i.e., fiber comprised the fermentation substrate of these bacteria either directly or indirectly. The fiber content in feces is an important environmental factor for these bacteria. The niche of bacteria varies, and increased fermentable substrates supply may promote colonization and fermentation of specific bacteria. Although these bacteria also correlated significantly with nutrient digestibility, it might be that feed intake and digestibility affected the fecal nutrient content and thus affected the abundance of these bacteria. Moreover, we found that the abundance of some bacteria did not correlate with fecal nutrient content, which is an important environmental factor, but correlated significantly and positively with the digestibility of the corresponding nutrients, such as
Enterobacteriaceae_
unclassified, Clostridium XlVb, and
Bifidobacterium. Although nutrient intake and digestion are major factors influencing the microbiota by influencing the types of substrates available in the digestive tract, some bacterial have the potential to interact directly with the host and affect nutrient digestion. However, the specific effects and mechanisms of these bacteria on nutrient digestion require further study.