In Silico Screening and Development of Microsatellite Markers for Genetic Analysis in Perca fluviatilis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
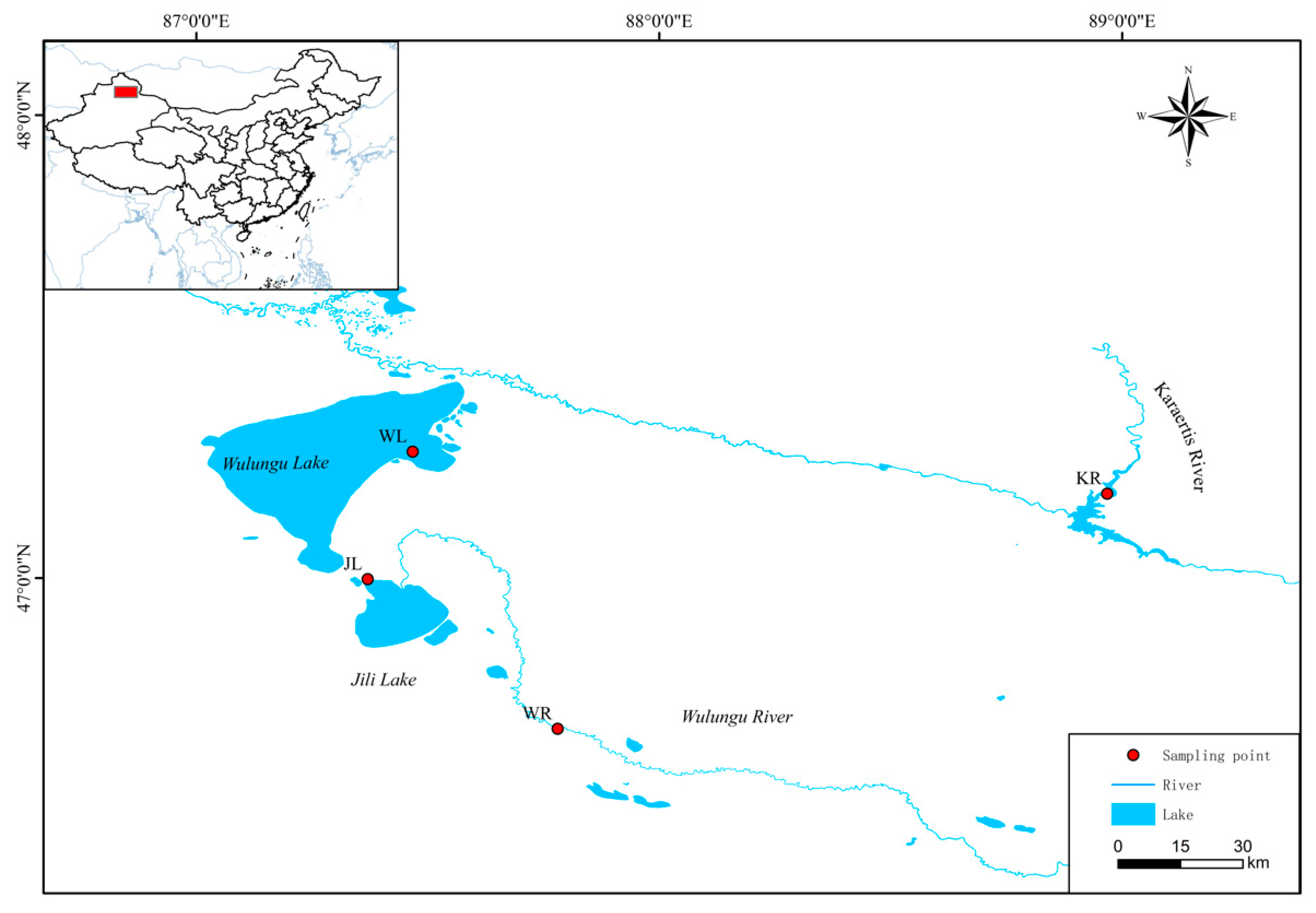
2.1. Sample Collection
2.2. Primer Design and Screening
2.3. DNA Extraction and PCR Amplification
2.4. Data Analysis
3. Results
3.1. Analysis of the Sequence Characteristics of Microsatellites in the P. fluviatilis Genome
3.2. Polymorphisms of the Microsatellite Markers
3.3. Genetic Diversity of P. fluviatilis Populations
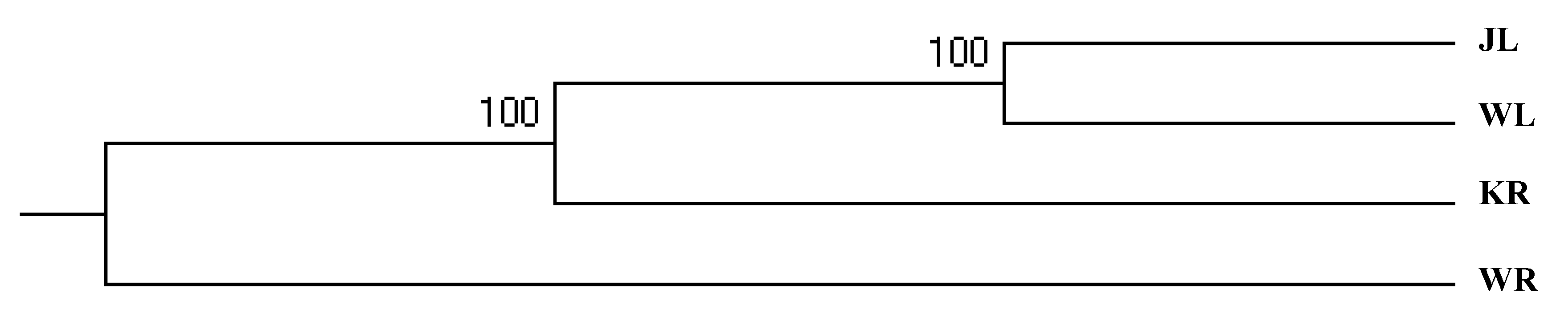
3.4. Genetic Differentiation of P. fluviatilis Populations
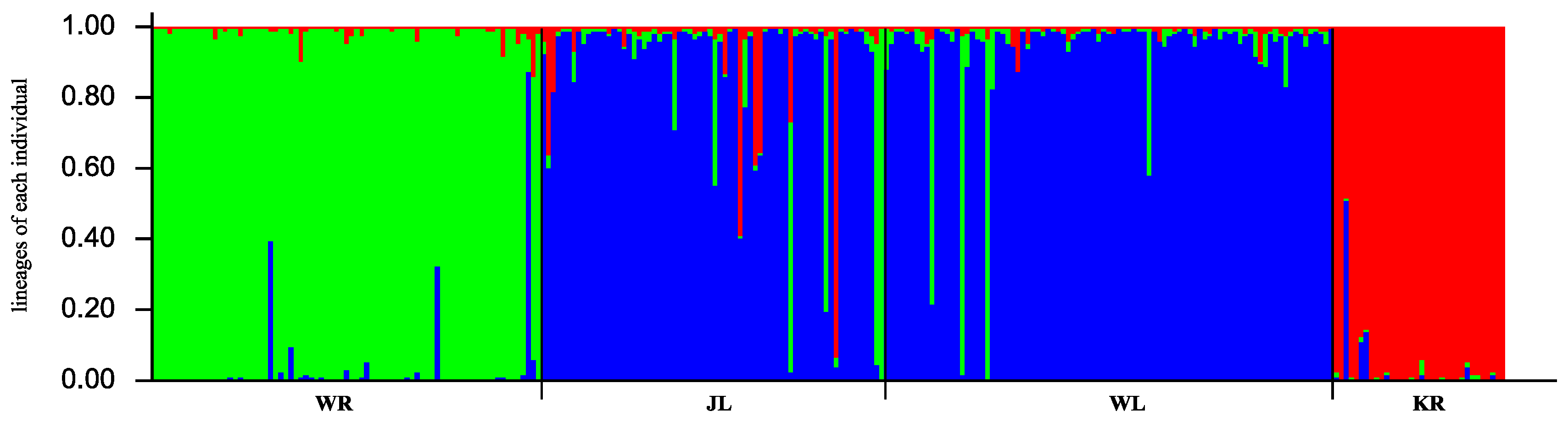
3.5. Analysis of the Population Genetic Components
4. Discussion
4.1. Characteristics and Screening of P. fluviatilis Microsatellite Sequences
4.2. Genetic Diversity and Differentiation of P. fluviatilis Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thorpe, J. Synopsis of biological data on the perch, Perca fluviatilis Linnaeus, 1785 and Perca flavescens Mitchill, 1814. FAO Fish. Synop. 2012, 113, 117–138. [Google Scholar]
- Fontaine, P. L’élevage de la perche commune, une voie de diversification pour l’aquaculture continentale. INRA Prod. Anim. 2004, 17, 189–193. [Google Scholar] [CrossRef]
- Hjelm, J.; Persson, L.; Christensen, B. Growth, morphological variation and ontogenetic niche shifts in perch (Perca fluviatilis) in relation to resource availability. Oecologia 2000, 122, 190–199. [Google Scholar] [CrossRef]
- Mairesse, G.; Thomas, M.; Gardeur, J.-N.; Brun-Bellut, J. Appearance and technological characteristics in wild and reared Eurasian perch, Perca fluviatilis (L.). Aquaculture 2004, 246, 295–311. [Google Scholar] [CrossRef]
- Stejskal, V.; Matoušek, J.; Prokešová, M.; Podhorec, P.; Křišťan, J.; Policar, T.; Gebauer, T. Fin damage and growth parameters relative to stocking density and feeding method in intensively cultured European perch (Perca fluviatilis L.). J. Fish Dis. 2020, 43, 253–262. [Google Scholar] [CrossRef]
- Sarosiek, B.; Dryl, K.; Kowalski, R.K.; Palińska-Żarska, K.; Żarski, D. Optimalisation of the Activation Medium and Effect of Inhibiting Activities of Acid Phosphatase, Lactate Dehydrogenase and β-N-Acetylglucosaminidase on the Fertilisation Success of Eurasian Perch (Perca fluviatilis L.). Animals 2022, 12, 307. [Google Scholar] [CrossRef]
- Qiao, D.; Ling, Q.; Yin, J.; Li, Y.; Cai, X.; He, Z. Development of embryo and yolk-sac stage larva of perch (Perca fluviatilis). J. Biol. 2006, 23, 34–38. [Google Scholar]
- Juhásová, Ľ.; Radačovská, A.; Bazsalovicsova, E.; Miklisová, D.; Bindzárová-Gereľová, M.; Králová-Hromadová, I. A study of the endohelminths of the European perch Perca fluviatilis L. from the central region of the Danube river basin in Slovakia. ZooKeys 2019, 899, 47. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.C.V.; Feld, C.K.; Hilt, S.; Manfrin, A.; Nachev, M.; Köster, D.; Jochmann, M.A.; Schmidt, T.C.; Sures, B.; Ziková, A. Eye fluke infection changes diet composition in juvenile European perch (Perca fluviatilis). Sci. Rep. 2021, 11, 1–14. [Google Scholar]
- Zare, M.; Tran, H.Q.; Prokešová, M.; Stejskal, V. Effects of Garlic Allium sativum Powder on Nutrient Digestibility, Haematology, and Immune and Stress Responses in Eurasian Perch Perca fluviatilis Juveniles. Animals 2021, 11, 2735. [Google Scholar] [CrossRef]
- Stejskal, V.; Tran, H.Q.; Prokesova, M.; Gebauer, T.; Giang, P.T.; Gai, F.; Gasco, L. Partially Defatted Hermetia illucens Larva Meal in Diet of Eurasian Perch (Perca fluviatilis) Juveniles. Animals 2020, 10, 1876. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, P.; Atkinson, R.; Gentile, G. Evaluating Genetic Diversity for the Conservation of the Threatened Galapagos Endemic Calandrinia galapagosa (Portulacaceae). Biotropica 2011, 43, 386–392. [Google Scholar] [CrossRef]
- Lu, C.; Sun, Z.; Xu, P.; Na, R.; Lv, W.; Cao, D.; Liu, T.; Zheng, X. Novel microsatellites reveal wild populations genetic variance in pike-perch (Sander lucioperca) in China. Aquac. Rep. 2022, 23, 101031. [Google Scholar] [CrossRef]
- Nesbø, C.L.; Fossheim, T.; Vøllestad, L.; Jakobsen, K. Genetic divergence and phylogeographic relationships among European perch (Perca fluviatilis) populations reflect glacial refugia and postglacial colonization. Mol. Ecol. 1999, 8, 1387–1404. [Google Scholar] [CrossRef] [PubMed]
- Schindler, D.E.; Hilborn, R.; Chasco, B.; Boatright, C.P.; Quinn, T.P.; Rogers, L.A.; Webster, M.S. Population diversity and the portfolio effect in an exploited species. Nature 2010, 465, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Hai, S.; Meng, W.; Yang, T.; Zhang, F. Genetic diversity analysis of Perca fluviatilis based on mtDNA control region sequences. Chin. J. Fish. 2012, 25, 38–41. [Google Scholar]
- Wu, F.; Hu, W.; Wang, C.; Guo, Y.; Mo, C. Genetic diverity of the cultivated and natural Perca fluviatilis. Acta Hydrobiol. Sin. 2016, 40, 181–188. [Google Scholar]
- Zhou, Y.; Wu, J.; Wang, Z.; Li, G.; Zhou, L.; Gui, J. Microsatellite polymorphism and genetic differentiation of different populations screened from genome survey sequencing in red-tail catfish (Hemibagrus wyckioides). Aquac. Rep. 2021, 19, 100614. [Google Scholar] [CrossRef]
- Fu, C.; Ai, Q.; Cai, L.; Qiu, F.; Yao, L.; Wu, H. Genetic Diversity and Population Dynamics of Leptobrachium leishanense (Anura: Megophryidae) as Determined by Tetranucleotide Microsatellite Markers Developed from Its Genome. Animals 2021, 11, 3560. [Google Scholar] [CrossRef]
- Khadher, S.B.; Agnèse, J.-F.; Milla, S.; Teletchea, F.; Fontaine, P. Patterns of genetic structure of Eurasian perch (Perca fluviatilis L.) in Lake Geneva at the end of the spawning season. J. Great Lakes Res. 2015, 41, 846–852. [Google Scholar] [CrossRef]
- Kánainé Sipos, D.; Csenki-Bakos, K.; Ősz, Á.; Bokor, Z.; Kotrik, L.; Żarski, D.; Ittzés, I.; Urbányi, B.; Kovács, B. Twelve new microsatellite loci of Eurasian perch Perca fluviatilis Linnaeus, 1758. Biol. Futura 2021, 72, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucl. Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, K.; Bana, N.; Bleier, N.; Sugár, L.; Stéger, V.; Nagy, J.; Wilhelm, J.; Kálmán, Z.; Barta, E.; Orosz, L.; et al. Mining the red deer genome (CerEla1.0) to develop X-and Y-chromosome-linked STR markers. PLoS ONE 2020, 15, e0242506. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, Z.; Li, W.; Zeng, T.; Zhang, X.; Li, J.; Zhang, W.; Yue, B. Isolation and strategies of novel tetranucleotide microsatellites with polymorphisms from different chromosomes of the rhesus monkey (Macaca mulatta). Mol. Biol. Rep. 2019, 46, 3955–3966. [Google Scholar] [CrossRef] [PubMed]
- Göl, Ş.; Göktay, M.; Allmer, J.; Doğanlar, S.; Frary, A. Newly developed SSR markers reveal genetic diversity and geographical clustering in spinach (Spinacia oleracea). Mol. Genet. Genomics 2017, 292, 847–855. [Google Scholar] [CrossRef]
- Li, S. Distribution Regionalization of Freshwater Fishes in China, 1st ed.; Science Press: Beijing, China, 1981; pp. 75–76. [Google Scholar]
- Ozerov, M.Y.; Ahmad, F.; Gross, R.; Pukk, L.; Kahar, S.; Kisand, V.; Vasemägi, A. Highly continuous genome assembly of Eurasian perch (Perca fluviatilis) using linked-read sequencing. G3 Genes Genomes Genet. 2018, 8, 3737–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Lu, C.; Zheng, X.; Cheng, L.; Xu, P.; Sun, X. A simple and effecient method for preserving fish fins. Chin. J. Fish. 2014, 27, 22–24. [Google Scholar]
- Becker, J.; Heun, M. Barley microsatellites: Allele variation and mapping. Plant Mol. Biol. 1995, 27, 835–845. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, K.M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Science Press: Beijing, China, 2002; pp. 463–471. [Google Scholar]
- Chi, T. Development and Application of Microsatellite Markers Based on Next-Generation Sequencing for Rana Dybowskii. Master’s Thesis, Shenyang Normal University, Shenyang, China, 2020. [Google Scholar]
- Yeh, F.; Yang, R.; Boyle, T.; Ye, Z.; Xiyan, J.; Yang, R.; Boyle, T. PopGene32, Microsoft Windows-Based Freeware for Population Genetic Analysis, Version 1.32; University of Alberta and Centre for International Forestry Research: Edmonton, AB, Canada, 1999.
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.Q.; Röder, M.; Sorrells, M.E. Frequencies and sequence characteristics of di-, tri-, and tetra-nucleotide microsatellites in wheat. Genome 1996, 39, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in Different Eukaryotic Genomes: Survey and Analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Ling, Q.; Li, C.; Wang, G.; Xu, Z.; Lu, G. Characterization of pikeperch (Sander lucioperca) transcriptome and development of SSR markers. Biochem. Syst. Ecol. 2016, 66, 188–195. [Google Scholar] [CrossRef]
- Gao, Z.; Luo, W.; Liu, H.; Zeng, C.; Liu, X.; Yi, S.; Wang, W. Transcriptome Analysis and SSR/SNP Markers Information of the Blunt Snout Bream (Megalobrama amblycephala). PLoS ONE 2012, 7, e42637. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Huang, H.; Liu, Y.; Wang, P.; Zhao, C.; Yan, L.; Qiao, X.; Qiu, L. Genome-wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). Aquac. Rep. 2021, 20, 100677. [Google Scholar]
- Cui, J.; Shen, X.; Yang, G.; Gong, Q.; Gu, Q. The Analysis of Simple Sequence Repeats in Takifugu rubripes Genome. J. Ocean Univ. China 2006, 36, 249–254, 272. [Google Scholar]
- Schorderet, D.F.; Gartler, S.M. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA 1992, 89, 957–961. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Mao, R.; Li, O.; Geng, L.; Sun, X.; Liang, L. Isolation and characterization of polymorphic tri- and tetra-nucleotide repeat microsatellite loci in common carp (Cyprinus carpio). Chin. J. Agric. Biotechol. 2009, 17, 979–987. [Google Scholar]
- Zheng, X.; Lu, C.; Sun, Z.; Dang, H.; Kuang, Y.; Cao, D.; Bai, Q.; Sun, X. Development of microsatellite markers based on transcriptome and population genetic structure analysis of introduced Salvelinus fontinalis. J. Fish. China 2020, 44, 1599–1608. [Google Scholar]
- Hong, K.; Guo, X.; Wang, Y.; Hou, Z.; Gu, Z.; Wang, A. Microsatellite markers screening from EST se-quences of Pinctada martensii (Dunker). J. Fish. China 2008, 32, 174–181. [Google Scholar]
- Barker, J.S.F. A global protocol for determining genetic distances among domestic livestock breeds. In Proceedings of the 5th World Congress on Genetics applied to Livestock Production, Guelph, ON, Canada, 7 August 1994. [Google Scholar]
- Zhou, L.; Xiao, Y.; Xia, W.; Yang, Y. Analysis of genetic diversity and population structure of oil palm (Elaeis guineensis) from China and Malaysia based on species-specific simple sequence repeat markers. Genet. Mol. Res. 2015, 14, 16247–16254. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv. Genet. 2006, 7, 167. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.R.Z. The Research of Breeding Regularity of Perca fluviatilis in Wulungu Lake. Master’s Thesis, Xinjiang Agricultural University, Xinjiang, China, 2015. [Google Scholar]
- Yang, X.; Qian, L.; Wu, H.; Fan, Z.; Wang, C. Population differentiation, bottleneck and selection of Eurasian perch (Perca fluviatilis L.) at the Asian edge of its natural range. Biochem. Syst. Ecol. 2012, 40, 6–12. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations, 1st ed.; University of Chicago Press: Chicago, IL, USA, 1984; p. 257. [Google Scholar]
- Xia, Q. “Yin E Ji Hai” outstanding achievement. Arid Environ. Monitor. 1989, 3, 11–12. [Google Scholar]
- Zuo, C.; Jiang, Z.; Li, S. Biology Studies of Cyprinus carpio in Lake Brento. Freshw. Fish. 2001, 31, 53–57. [Google Scholar]

| Locus | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) | Repeat Motif | Fragment Size/bp |
|---|---|---|---|---|
| HLJHL007 | GCTAATGCCTGACCACCACT | CAGGTCCCTGGAGAAGAGAGA | (ACAG)10 | 109–130 |
| HLJHL022 | CCCTGAGTGGAGCACATACC | GGCCGAATTTTGCCTTTGTA | (ACAG)12 | 100–131 |
| HLJHL045 | GATGGTCTCACGAGGCTAGC | TCGGTTTGGATGGTCGGTTT | (ACAG)14 | 86–114 |
| HLJHL052 | ACCTGAAGGCAAGTGGATGG | CCTGGCCCTGAATGTACCAA | (ACAG)15 | 92–120 |
| HLJHL056 | CAGCAGCCTGATCAGCATCA | TGCTTTACTTCCTCATCACAGC | (ACAG)15 | 120–148 |
| HLJHL059 | GGTTTCAAGGGAGAGGGAGG | GCAGCTCATGAACAACTCTGC | (ACAG)15 | 92–145 |
| HLJHL084 | AGCTCGACTAGGGTGACTGT | CCGAGCACTGCTACGAAACT | (ACAG)18 | 143–174 |
| HLJHL089 | ATTCACACAAACAAGCAGGC | GCTATGTGGCTCTTTGTATGCC | (ACAG)19 | 100–160 |
| HLJHL090 | GGCGCTGTCCATGGTACTAA | ACCACGAAAAGCAAGAAGGC | (ACAG)19 | 117–162 |
| HLJHL094 | ACTCACCATACGCATGTGCT | CTCCTCAAAGTCGCCTTCCA | (ACAG)19 | 114–186 |
| HLJHL104 | CCCAAATTTCCTGACAACCCA | GGACTGTCCCGTGTTTCTCA | (AGAT)10 | 132–223 |
| HLJHL105 | GCGCGATAAAATAATTGTCGGC | TCAGGCTCAGGAATTTCTTTCA | (AGAT)10 | 107–148 |
| HLJHL107 | AGACAGGGTGATAGTTACATCCA | TGTGCAAAATTTACATGGGATGA | (AGAT)10 | 118–188 |
| HLJHL121 | ATTGGCATCAGAGCAAGCTG | ATGGGGCTTTGACGTGAGAG | (AGAT)11 | 184–265 |
| HLJHL152 | GCCAACACCCTATAACTGAAGC | CGTTGTTGCCAATGGAATGC | (AGAT)14 | 140–185 |
| HLJHL153 | GGCTGATAAACATAGGCCTATGC | AGCTACTGGGATCTTGAAGGT | (AGAT)14 | 123–151 |
| HLJHL164 | CCACCTCTGCCACCTCATAC | CGAAGGGATCTCCATCTGCT | (AGAT)15 | 104–175 |
| HLJHL165 | TCCAGTTGTCACTTCAGCGT | AGGACACATTTCCTTCGGGA | (AGAT)15 | 88–194 |
| HLJHL167 | CGTTTTGGATATGTGCCATGT | TGGCACATATCTAAAACGTGGT | (AGAT)15 | 140–188 |
| HLJHL169 | GCAGGGGCAAACAGTCATCA | TCTGTGAGCTACTGGGACCT | (AGAT)16 | 100–152 |
| HLJHL172 | ACAGCCCATAACACAGCAGT | TCTGCATGAACTAAAGTGTGACA | (AGAT)16 | 126–187 |
| HLJHL174 | TCAGCTGCGGATTATTACACA | GCAGTGATATTGCAACAGGAAA | (AGAT)16 | 131–162 |
| HLJHL179 | GGTGATACATAGATAGGTAGGTCGG | TCTGGTAGTCTCAGCTCGCT | (AGAT)16 | 96–153 |
| HLJHL183 | TGTTGTCAGTGTGTTCATCCA | TGCATGGTGTTTTAAGTCAGGG | (AGAT)17 | 125–168 |
| HLJHL186 | CAACCAGCTTCAACCCGTTG | TCCACCTCTCCCTTTCCCTT | (AGAT)17 | 123–237 |
| HLJHL189 | CCTCCTGTGTTTTGTGTCTTGG | TCTCCAGTACTCACAATGGCT | (AGAT)18 | 124–175 |
| HLJHL192 | TGGTTCTACAAGCTGCCTAAA | AACCAGGCGTTGAGTTTCAA | (AGAT)18 | 102–168 |
| HLJHL196 | TCTGAGACAAAGGGACATGAAT | CAGGAATTTCCCCAGTGTGG | (AGAT)19 | 111–183 |
| HLJHL199 | TGGACTAAGACTGCCTACTGC | CCTTGAGTTCACTTGCGTGT | (AGAT)20 | 134–190 |
| Repeat Motif Type | Count | Proportion |
|---|---|---|
| (AGAT)n | 1556 | 17.78% |
| (ACAG)n | 1339 | 15.30% |
| (ATCC)n | 752 | 8.59% |
| (AAAT)n | 645 | 7.37% |
| (AAAG)n | 610 | 6.97% |
| (ACAT)n | 560 | 6.40% |
| (AATC)n | 485 | 5.54% |
| (ACGC)n | 442 | 5.05% |
| (AGGG)n | 430 | 4.91% |
| (AATG)n | 302 | 3.45% |
| (AAGG)n | 234 | 2.67% |
| (AAAC)n | 221 | 2.53% |
| (AAGT)n | 201 | 2.30% |
| (ACTC)n | 173 | 1.98% |
| (ACTG)n | 135 | 1.54% |
| (AACT)n | 127 | 1.45% |
| (AGGC)n | 107 | 1.22% |
| (AGCT)n | 101 | 1.15% |
| (AGCC)n | 75 | 0.86% |
| (AATT)n | 69 | 0.79% |
| (AACC)n | 52 | 0.59% |
| (ACCT)n | 40 | 0.46% |
| (ACCC)n | 37 | 0.42% |
| (ATGC)n | 22 | 0.25% |
| (ACCG)n | 9 | 0.10% |
| (AAGC)n | 8 | 0.09% |
| (AGCG)n | 7 | 0.08% |
| (ATCG)n | 4 | 0.05% |
| (ACGG)n | 3 | 0.03% |
| (CCCG)n | 2 | 0.02% |
| (ACGT)n | 2 | 0.02% |
| (CCGG)n | 1 | 0.01% |
| Locus | Na | Ne | Ho | He | PIC |
|---|---|---|---|---|---|
| HLJHL007 | 4 | 2.201 | 0.403 | 0.547 | 0.489 |
| HLJHL022 | 6 | 2.169 | 0.496 | 0.540 | 0.450 |
| HLJHL045 | 8 | 2.133 | 0.485 | 0.532 | 0.496 |
| HLJHL052 | 4 | 1.784 | 0.425 | 0.440 | 0.364 |
| HLJHL056 | 5 | 2.827 | 0.623 | 0.647 | 0.578 |
| HLJHL059 | 10 | 4.444 | 0.675 | 0.776 | 0.741 |
| HLJHL084 | 9 | 1.310 | 0.239 | 0.237 | 0.225 |
| HLJHL089 | 11 | 2.264 | 0.489 | 0.559 | 0.528 |
| HLJHL090 | 11 | 1.671 | 0.373 | 0.402 | 0.389 |
| HLJHL094 | 18 | 3.057 | 0.623 | 0.674 | 0.661 |
| HLJHL104 | 17 | 7.506 | 0.869 | 0.868 | 0.853 |
| HLJHL105 | 11 | 7.389 | 0.847 | 0.866 | 0.850 |
| HLJHL107 | 18 | 9.968 | 0.896 | 0.901 | 0.892 |
| HLJHL121 | 19 | 10.077 | 0.877 | 0.902 | 0.893 |
| HLJHL152 | 13 | 5.949 | 0.791 | 0.833 | 0.811 |
| HLJHL153 | 8 | 4.181 | 0.716 | 0.762 | 0.725 |
| HLJHL164 | 14 | 5.863 | 0.776 | 0.831 | 0.809 |
| HLJHL165 | 20 | 3.269 | 0.608 | 0.695 | 0.650 |
| HLJHL167 | 13 | 6.486 | 0.519 | 0.847 | 0.827 |
| HLJHL169 | 9 | 3.256 | 0.690 | 0.694 | 0.641 |
| HLJHL172 | 15 | 4.677 | 0.750 | 0.788 | 0.765 |
| HLJHL174 | 9 | 4.714 | 0.731 | 0.789 | 0.760 |
| HLJHL179 | 11 | 3.273 | 0.612 | 0.696 | 0.658 |
| HLJHL183 | 12 | 3.973 | 0.698 | 0.750 | 0.720 |
| HLJHL186 | 29 | 10.914 | 0.694 | 0.910 | 0.901 |
| HLJHL192 | 14 | 5.783 | 0.694 | 0.829 | 0.808 |
| HLJHL196 | 15 | 2.389 | 0.526 | 0.583 | 0.512 |
| HLJHL199 | 16 | 9.361 | 0.843 | 0.895 | 0.884 |
| HLJHL200 | 15 | 6.564 | 0.813 | 0.849 | 0.833 |
| Source of Variation | Degree of Freedom (df) | Sum of Squares | Variance Components | Percentage of Variation/% |
|---|---|---|---|---|
| Among populations | 3 | 334.462 | 0.784 | 7.44 |
| Within populations | 532 | 5188.229 | 9.752 | 92.56 |
| Total | 535 | 5522.690 | 10.536 |
| Population | WR | JL | WL | KR |
|---|---|---|---|---|
| WR | / | 0.087 * | 0.071 * | 0.158 * |
| JL | 0.205 | / | 0.003 | 0.116 * |
| WL | 0.157 | 0.027 | / | 0.100 * |
| KR | 0.288 | 0.244 | 0.1987 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Lu, C.; Sun, Z.; Kuang, Y.; Cao, D.; Huo, T.; Li, C.; Jin, H.; Zheng, X. In Silico Screening and Development of Microsatellite Markers for Genetic Analysis in Perca fluviatilis. Animals 2022, 12, 1809. https://doi.org/10.3390/ani12141809
Xu P, Lu C, Sun Z, Kuang Y, Cao D, Huo T, Li C, Jin H, Zheng X. In Silico Screening and Development of Microsatellite Markers for Genetic Analysis in Perca fluviatilis. Animals. 2022; 12(14):1809. https://doi.org/10.3390/ani12141809
Chicago/Turabian StyleXu, Pu, Cuiyun Lu, Zhipeng Sun, Youyi Kuang, Dingchen Cao, Tangbin Huo, Chao Li, Hongyu Jin, and Xianhu Zheng. 2022. "In Silico Screening and Development of Microsatellite Markers for Genetic Analysis in Perca fluviatilis" Animals 12, no. 14: 1809. https://doi.org/10.3390/ani12141809
APA StyleXu, P., Lu, C., Sun, Z., Kuang, Y., Cao, D., Huo, T., Li, C., Jin, H., & Zheng, X. (2022). In Silico Screening and Development of Microsatellite Markers for Genetic Analysis in Perca fluviatilis. Animals, 12(14), 1809. https://doi.org/10.3390/ani12141809






