Screening of Microbial Fermentation Products for Anti-M. tuberculosis Activity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Medium
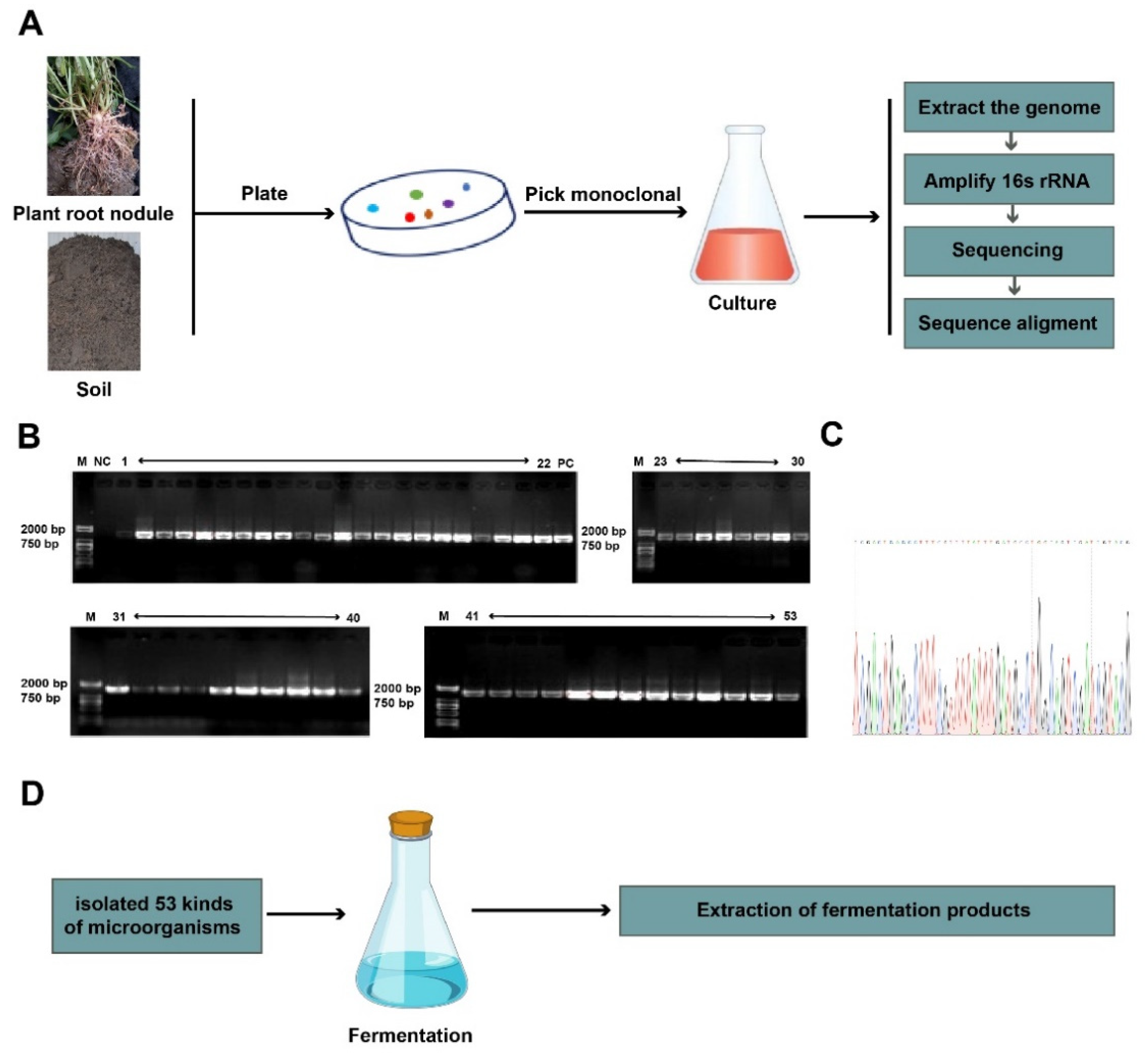
2.2. Isolation and Identification of Soil Microorganisms
2.3. Extraction of Fermentation Products
2.4. Bacterial Strain and Cell Cultures
2.5. Microplate Alamar Blue Assay
2.6. Determination of the Minimum Inhibitory Concentration of Selected Fermentation Products
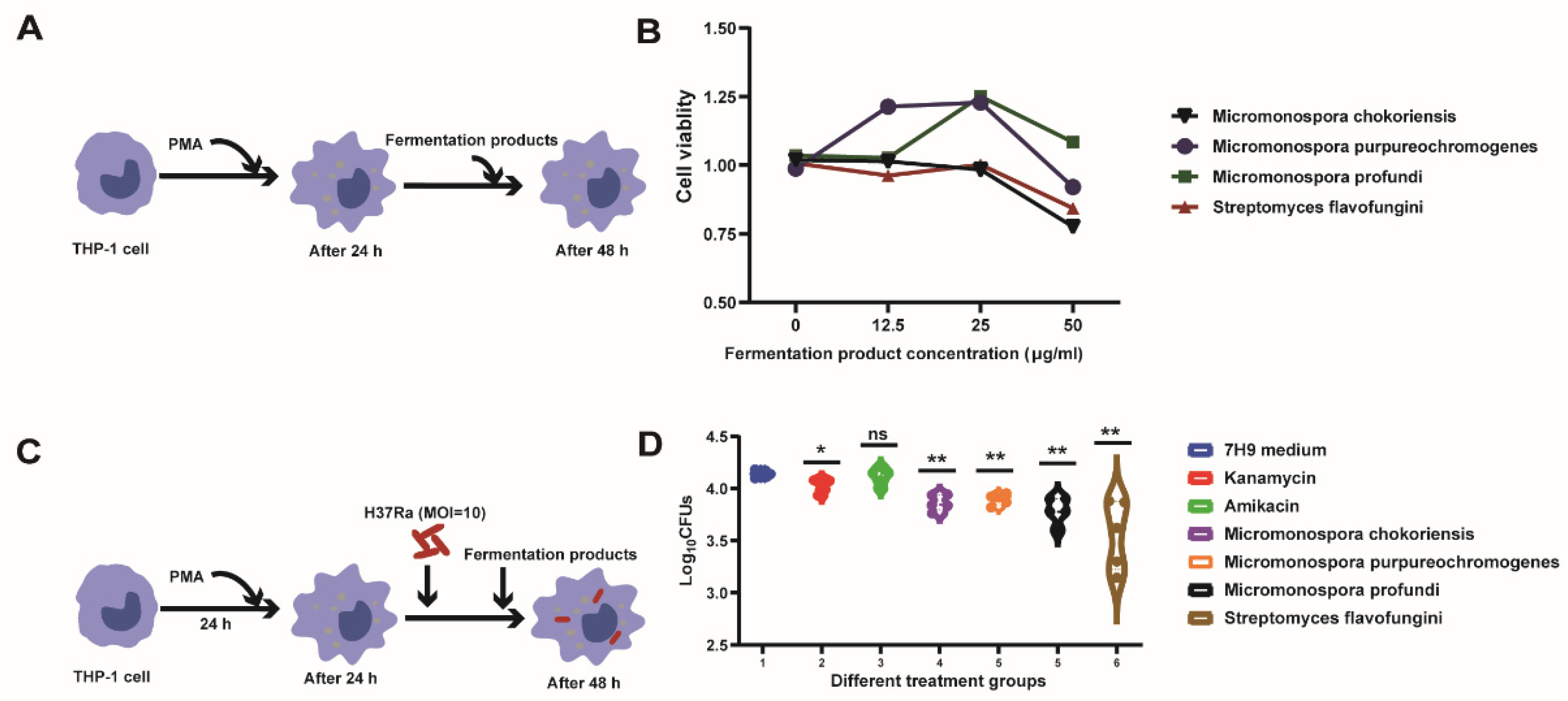
2.7. Cell Viability Analysis
2.8. Drug Susceptibility Testing against Intracellular M.tb
2.9. Isolated Total RNA and qRT-PCR
2.10. Statistical Analysis
3. Results
3.1. Total of 53 Fermentation Products Were Isolated from the Chickpea Root Nodules and Kaline Soil
3.2. Screening the Fermented Products against M.tb In Vitro
3.3. Intracellular Bactericidal Effects of Fermentation Products
3.4. Effect of Fermentation Products on M.tb’s Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Zumla, A.; Nahid, P.; Cole, S.T. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 2013, 12, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Tao, N.N.; He, X.C.; Zhang, X.X.; Liu, Y.; Yu, C.B.; Li, H.C. Drug-Resistant Tuberculosis among Children, China, 2006–2015. Emerg. Infect. Dis. 2017, 23, 1800–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, J.; Marais, B.J. Multidrug-resistant tuberculosis infection and disease in children: A review of new and repurposed drugs. Ther. Adv. Infect. Dis. 2019, 6, 2049936119864737. [Google Scholar] [CrossRef] [PubMed]
- Koul, A.; Arnoult, E.; Lounis, N.; Guillemont, J.; Andries, K. The challenge of new drug discovery for tuberculosis. Nature 2011, 469, 483–490. [Google Scholar] [CrossRef]
- Arthur, P.K.; Amarh, V.; Cramer, P.; Arkaifie, G.B.; Blessie, E.J.S.; Fuseini, M.S.; Carilo, I.; Yeboah, R.; Asare, L.; Robertson, B.D. Characterization of Two New Multidrug-Resistant Strains of Mycobacterium smegmatis: Tools for Routine In Vitro Screening of Novel Anti-Mycobacterial Agents. Antibiotics 2019, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [Green Version]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Gohlmann, H.W.H.; Neefs, J.M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Guzman, J.D.; Gupta, A.; Bucar, F.; Gibbons, S.; Bhakta, S. Antimycobacterials from natural sources: Ancient times, antibiotic era and novel scaffolds. Front. Biosci.-Landmark 2012, 17, 1861–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, M. Development of new antituberculosis drugs from natural products. Biosci. Biotechnol. Biochem. 2017, 81, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.W.; Zhou, X.F.; Lin, X.P.; Qin, X.C.; Zhang, T.Y.; Wang, J.F.; Tu, Z.C.; Yang, B.; Liao, S.R.; Tian, Y.Q.; et al. Antituberculosis compounds from a deep-sea-derived fungus Aspergillus sp. SCSIO Ind09F01. Nat. Prod. Res. 2017, 31, 1958–1962. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Ishizaki, Y.; Takahashi, Y. New antituberculous drugs derived from natural products: Current perspectives and issues in antituberculous drug development. J. Antibiot. 2018, 71, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Elad, N.; Baron, S.; Peleg, Y.; Albeck, S.; Grunwald, J.; Raviv, G.; Shakked, Z.; Zimhony, O.; Diskin, R. Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution. Nat. Commun. 2018, 9, 3886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kassama, Y.; Young, M.; Kell, D.B.; Goodacre, R. Differentiation of Micromonospora isolates from a coastal sediment in Wales on the basis of Fourier transform infrared spectroscopy, 16S rRNA sequence analysis, and the amplified fragment length polymorphism technique. Appl. Environ. Microbiol. 2004, 70, 6619–6627. [Google Scholar] [CrossRef] [Green Version]
- Talukdar, M.; Bordoloi, M.; Dutta, P.P.; Saikia, S.; Kolita, B.; Talukdar, S.; Nath, S.; Yadav, A.; Saikia, R.; Jha, D.K.; et al. Structure elucidation and biological activity of antibacterial compound from Micromonospora auratinigra, a soil Actinomycetes. J. Appl. Microbiol. 2016, 121, 973–987. [Google Scholar] [CrossRef] [Green Version]
- Tiffin, A.I. Flavofungin, a New Crystalline Antifungal Antibiotic Origin and Biological Properties. Nature 1958, 181, 908–909. [Google Scholar]
- Mishra, S.K.; Tripathi, G.; Kishore, N.; Singh, R.K.; Singh, A.; Tiwari, V.K. Drug development against tuberculosis: Impact of alkaloids. Eur. J. Med. Chem. 2017, 137, 504–544. [Google Scholar] [CrossRef] [PubMed]
- Salaemae, W.; Azhar, A.; Booker, G.W.; Polyak, S.W. Biotin biosynthesis in Mycobacterium tuberculosis: Physiology, biochemistry and molecular intervention. Protein Cell 2011, 2, 691–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bockman, M.R.; Engelhart, C.A.; Cramer, J.D.; Howe, M.D.; Mishra, N.K.; Zimmerman, M.; Larson, P.; Alvarez-Cabrera, N.; Park, S.W.; Boshoff, H.I.M.; et al. Investigation of (S)-(-)-Acidomycin: A Selective Antimycobacterial Natural Product That Inhibits Biotin Synthase. ACS Infect. Dis. 2019, 5, 598–617. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Rao, S.P.; Noble, C.G. Structural basis of mycobacterial inhibition by cyclomarin A. J. Biol. Chem. 2013, 288, 30883–30891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Cao, X.; Lei, Y.; Reheman, A.; Zhou, W.; Yang, B.; Zhang, W.; Xu, W.; Dong, S.; Tyagi, R.; et al. Distinct Persistence Fate of Mycobacterium tuberculosis in Various Types of Cells. mSystems 2021, 6, e00783-21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, B.; Zou, Y.Y.; Rahman, K.; Cao, X.J.; Lei, Y.Y.; Lai, R.; Fu, Z.F.; Chen, X.; Cao, G. Screening of Compounds for Anti-tuberculosis Activity, and in vitro and in vivo Evaluation of Potential Candidates. Front. Microbiol. 2021, 12, 1619. [Google Scholar] [CrossRef] [PubMed]
- Rego, A.M.; Alves da Silva, D.; Ferreira, N.V.; de Pina, L.C.; Evaristo, J.A.M.; Caprini Evaristo, G.P.; Nogueira, F.C.S.; Ochs, S.M.; Amaral, J.J.; Ferreira, R.B.R.; et al. Metabolic profiles of multidrug resistant and extensively drug resistant Mycobacterium tuberculosis unveiled by metabolomics. Tuberculosis 2021, 126, 102043. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Quan, D.; Nagalingam, G.; Payne, R.; Triccas, J.A. New tuberculosis drug leads from naturally occurring compounds. Int. J. Infect. Dis. 2017, 56, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.; Bhattarcharya, D.; Saha, M.; Mukherjee, J.; Karmakar, S. Evaluation of antimicrobial activity of the extract of Streptomyces euryhalinus isolated from the Indian Sundarbans. Arch. Microbiol. 2022, 204, 34. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Liu, X.T.; Zhang, L.X.; Quinn, R.J.; Feng, Y.J. Anti-mycobacterial natural products and mechanisms of action. Nat. Prod. Rep. 2022, 39, 77–89. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, M.A.; Gerrick, E.R.; Xu, W.Z.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive Essentiality Analysis of the Mycobacterium tuberculosis Genome via Saturating Transposon Mutagenesis. Mbio 2017, 8, e02133-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, J.E.; Gawronski, J.D.; DeJesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef] [Green Version]
- Minato, Y.; Gohl, D.M.; Thiede, J.M.; Chacon, J.M.; Harcombe, W.R.; Maruyama, F.; Baughn, A.D. Genomewide Assessment of Mycobacterium tuberculosis Conditionally Essential Metabolic Pathways. MSystems 2019, 4, e00070-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckert, P.; Hillemann, D.; Kohl, T.A.; Kalinowski, J.; Richter, E.; Niemann, S.; Feuerriegel, S. rplC T460C Identified as a Dominant Mutation in Linezolid-Resistant Mycobacterium tuberculosis Strains. Antimicrob. Agents Chemother. 2012, 56, 2743–2745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smittipat, N.; Juthayothin, T.; Billamas, P.; Jaitrong, S.; Rukseree, K.; Dokladda, K.; Chaiyasirinroje, B.; Disratthakit, A.; Chaiprasert, A.; Mahasirimongkol, S.; et al. Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J. Glob. Antimicrob. Resist. 2016, 4, 5–10. [Google Scholar] [CrossRef]
- Wolf, N.M.; Lee, H.; Choules, M.P.; Pauli, G.F.; Phansalkar, R.; Anderson, J.R.; Gao, W.; Ren, J.; Santarsiero, B.D.; Lee, H.; et al. High-Resolution Structure of ClpC1-Rufomycin and Ligand Binding Studies Provide a Framework to Design and Optimize Anti-Tuberculosis Leads. ACS Infect. Dis. 2019, 5, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Loewen, P.C.; De Silva, P.M.; Donald, L.J.; Switala, J.; Villanueva, J.; Fita, I.; Kumar, A. KatG-Mediated Oxidation Leading to Reduced Susceptibility of Bacteria to Kanamycin. ACS Omega 2018, 3, 4213–4219. [Google Scholar] [CrossRef] [Green Version]
- Annaraj, P.D.; Kadirvel, P.; Subramanian, A.; Anishetty, S. Free enzyme dynamics of CmaA3 and CmaA2 cyclopropane mycolic acid synthases from Mycobacterium tuberculosis: Insights into residues with potential significance in cyclopropanation. J. Mol. Graph. Model. 2019, 91, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, N.; Bhatti, S.; Abbas, S.N.; Shahid, S.; Nasir, S.B. The pncA gene mutations of Mycobacterium tuberculosis in multidrug-resistant tuberculosis. Biotechnol. Appl. Bioc. 2021, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]


| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| clpC1 | 5′-GTCGTCCTGGCTCAGGAAGA-3′ | 5′-GACCTGACTGCGCACACCTT-3′ |
| katG | 5′-TACAGAAACCACCACCGGAG-3′ | 5′-TAGTCGAACGCCGCACCCAT-3′ |
| pncA | 5′-GACGTGCAGAACGACTTCTG-3′ | 5′-ATAGTCCGGTGTGCCGGAGA-3′ |
| cmaA2 | 5′-ACAAGCGGCACGCAGCTCAA-3′ | 5′-TACTGCGCCTCTTCCAGCGT-3′ |
| rpoB | 5′-ACAGCCGCTAGTCCTAGTCC-3′ | 5′-CGAACCGATCAGCCACTCGA-3′ |
| rpsL | 5′-TCGGGACAAGATCAGTAAGG-3′ | 5′-ATGTACGCCGTGACCTCGAC-3′ |
| rplC | 5′-TATGACGCAGGTATTCGACG-3′ | 5′-GACCTTGCGTGGGCTGATCT-3′ |
| bioB | 5′-ATGGCGGGAACAACTCGGAC-3′ | 5′-TGATGATGCCTTCGACCTCG-3′ |
| 16S rDNA | 5′-AGAGTTTGATCCTGGCTC-3′ | 5′-CGGCTACCTTGTTACGACTT-3′ |
| Num. | Microorganisms | Num. | Microorganisms |
|---|---|---|---|
| 1 | Micromonospora taraxaci | 28 | Nonomuraea kuesteri |
| 2 | Micromonospora chokoriensis | 29 | Streptomyces flavofungini |
| 3 | Micromonospora purpureochromogenes | 30 | Streptomyces griseoincarnatus |
| 4 | Micromonospora halophytica | 31 | Actinoplanes alaerensis |
| 5 | Micromonospora lutea | 32 | Streptomyces mangrovi |
| 6 | Micromonospora saelicesensis | 33 | Streptomyces griseus subsp. Griseus |
| 7 | Micromonospora inaquosa | 34 | Amycolatopsis roodepoortensis |
| 8 | Micromonospora vinacae | 35 | Streptomyces flavofungini |
| 9 | Micromonospora zamorensis | 36 | Lentzea jiangxiensis |
| 10 | Micromonospora chalea | 37 | Amycolatopsis lurida |
| 11 | Micromonospora terminaliae | 38 | Amycolatopsis lurida-1 |
| 12 | Micromonospora profundi | 39 | Nonomuraea harbinensis |
| 13 | Micromonospora spongocola | 40 | Micromonospora sediminimaris |
| 14 | Micromonospora auratinigra | 41 | Actinoplanes alaerensis |
| 15 | Micromonospora cremea | 42 | Micromonospora andamanensis |
| 16 | Micromonospora gifhorensis | 43 | Actinoplanes |
| 17 | Micromonospora phytophia | 44 | Actinoplanes xinjiangensis |
| 18 | Micromonospora andamanensis | 45 | Actinoplanes utahensis |
| 19 | Micromonospora fiedleri | 46 | Actinoplanes brasiliensis |
| 20 | Micromonospora soli | 47 | Actinoplanes rectilineatus |
| 21 | Micromonospora oryzae | 48 | Actinoplanes cyaneus |
| 22 | Micromonospora dersiti | 49 | Actinoplanes atraurantiacus |
| 23 | Micromonospora inositola | 50 | Actinoplanes lutulentus |
| 24 | Amycolatopsis marina | 51 | Actinoplanes abujensis |
| 25 | Streptomyces formicae | 52 | Actinoplanes missouriensis |
| 26 | Nocardiopsis terrae | 53 | Actinoplanes abujensis |
| 27 | Streptomyces griseorubens |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reheman, A.; Lu, D.; Wang, Y.; Chen, X.; Cao, G.; Wan, C. Screening of Microbial Fermentation Products for Anti-M. tuberculosis Activity. Animals 2022, 12, 1947. https://doi.org/10.3390/ani12151947
Reheman A, Lu D, Wang Y, Chen X, Cao G, Wan C. Screening of Microbial Fermentation Products for Anti-M. tuberculosis Activity. Animals. 2022; 12(15):1947. https://doi.org/10.3390/ani12151947
Chicago/Turabian StyleReheman, Aikebaier, Di Lu, Yifan Wang, Xi Chen, Gang Cao, and Chuanxing Wan. 2022. "Screening of Microbial Fermentation Products for Anti-M. tuberculosis Activity" Animals 12, no. 15: 1947. https://doi.org/10.3390/ani12151947





