Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3Cpro
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Andrographolides
2.3. Cytotoxicity Assay
2.4. Antiviral Activities of the Andrographolides
2.5. Immunoperoxidase Monolayer Assay (IPMA)
2.6. FMDV RNA Quantification Using RT-qPCR
2.7. Intracellular Protease Inhibitory Assay
2.8. Optimization of Interferon (IFN) β Treatment in HEK 293T Cells
2.9. Evaluating Effects of Andrographolides on the Inhibition of Interferon Stimulating Gene (ISG) Expressions by IFN-Antagonist Activity of 3Cpro
2.10. Molecular Docking and Protein-Ligand Interaction
3. Results
3.1. Cytotoxicity of the Andrographolides on BHK-21 and HEK 293T Cells
3.2. Antiviral Activities of AGL and DAG after Infection
3.3. No Direct Effect of AGL and DAG on Extracellular Viruses
3.4. Effects of AGL and DAG on FMDV 3Cpro
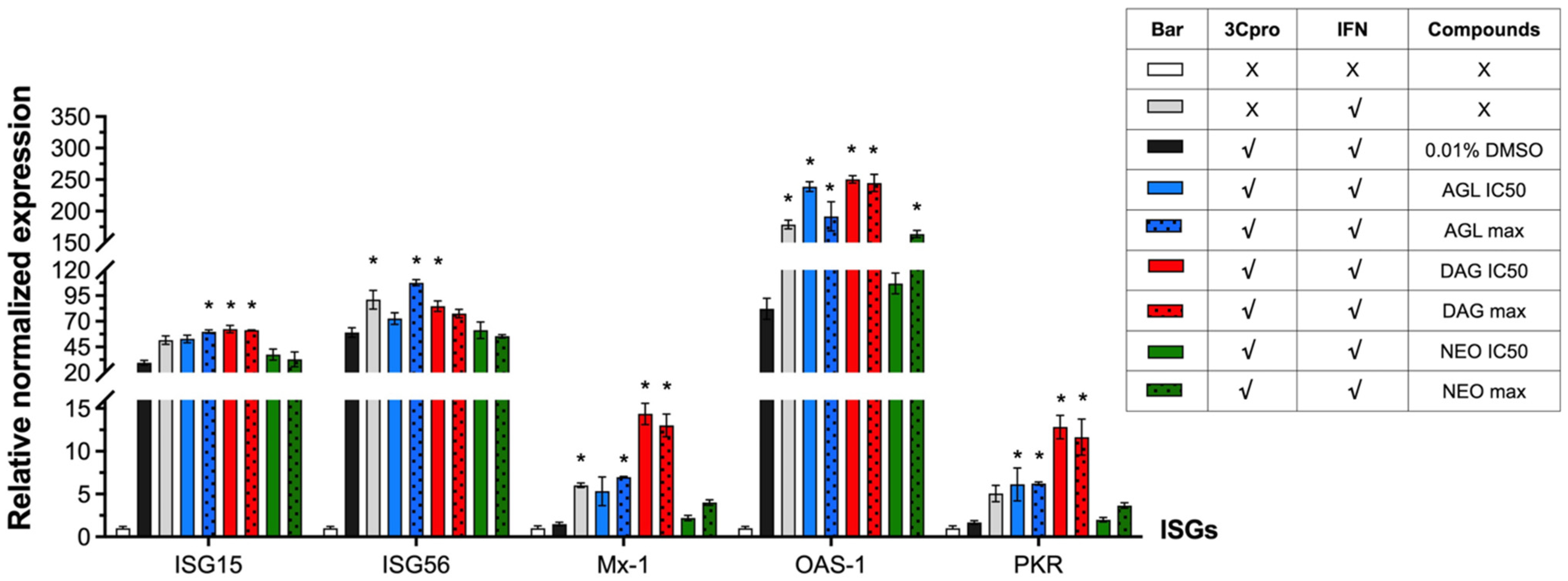
3.5. Interfering IFN-Antagonist Activity of FMDV 3Cpro by Andrographolides
3.6. Binding Preferences among Andrographolides and FMDV 3pro Active Residues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamal, S.M.; Belsham, G.J. Foot-and-mouth disease: Past, present and future. Vet. Res. 2013, 44, 116. [Google Scholar] [CrossRef]
- The Food and Agriculture Organization (FAO). Food-and-Mouth Disease, April–June 2021, Quarterly Report. FAST Reports: Foot-and-Mouth and Similar Transboundary Animal Diseases. 2021. Available online: http://www.fao.org/3/cb5998en/cb5998en.pdf. (accessed on 21 January 2022).
- Paton, D.J.; Di Nardo, A.; Knowles, N.J.; Wadsworth, J.; Pituco, E.M.; Cosivi, O.; Rivera, A.M.; Kassimi, L.B.; Brocchi, E.; de Clercq, K.; et al. The history of foot-and-mouth disease virus serotype C: The first known extinct serotype? Virus Evol. 2021, 7, veab009. [Google Scholar] [CrossRef] [PubMed]
- Belsham, G.J. Towards improvements in foot-and-mouth disease vaccine performance. Acta Vet. Scand. 2020, 62, 20. [Google Scholar] [CrossRef] [PubMed]
- Fry, E.E.; Stuart, D.I.; Rowlands, D.J. The structure of foot-and-mouth disease virus. Curr. Top. Microbiol. Immunol. 2005, 288, 71–101. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.W.; Grubman, M.J.; Baxt, B. Molecular basis of pathogenesis of FMDV. Virus Res. 2003, 91, 9–32. [Google Scholar] [CrossRef]
- Carrillo, C.; Tulman, E.R.; Delhon, G.; Lu, Z.; Carreno, A.; Vagnozzi, A.; Kutish, G.F.; Rock, D.L. Comparative genomics of foot-and-mouth disease virus. J. Virol. 2005, 79, 6487–6504. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Order Picornavirales. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 835–839. [Google Scholar] [CrossRef]
- Thomas, E.; Stewart, L.E.; Darley, B.A.; Pham, A.M.; Esteban, I.; Panda, S.S. Plant-based natural products and extracts: Potential source to develop new antiviral drug candidates. Molecules 2021, 26, 6197. [Google Scholar] [CrossRef]
- Attia, Y.A.; Alagawany, M.M.; Farag, M.R.; Alkhatib, F.M.; Khafaga, A.F.; Abdel-Moneim, A.-M.E.; Asiry, K.A.; Mesalam, N.M.; Shafi, M.E.; Al-Harthi, M.A.; et al. Phytogenic products and phytochemicals as a candidate strategy to improve tolerance to coronavirus. Front. Vet. Sci. 2020, 7, 573159. [Google Scholar] [CrossRef]
- Jayakumar, T.; Hsieh, C.Y.; Lee, J.J.; Sheu, J.R. Experimental and clinical pharmacology of Andrographis paniculata and its major bioactive phytoconstituent andrographolide. Evid.-Based Complement. Altern. Med. 2013, 2013, 846740. [Google Scholar] [CrossRef]
- Gupta, S.; Mishra, K.P.; Ganju, L. Broad-spectrum antiviral properties of andrographolide. Arch. Virol. 2017, 162, 611–623. [Google Scholar] [CrossRef]
- Burgos, R.A.; Alarcón, P.; Quiroga, J.; Manosalva, C.; Hancke, J. Andrographolide, an anti-inflammatory multitarget drug: All roads lead to cellular metabolism. Molecules 2020, 26, 5. [Google Scholar] [CrossRef] [PubMed]
- Pholphana, N.; Rangkadilok, N.; Saehun, J.; Ritruechai, S.; Satayavivad, J. Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm. f.) Nees (Chuanxinlian). Chin. Med. 2013, 8, 2. [Google Scholar] [CrossRef]
- Wang, D.; Guo, H.; Chang, J.; Wang, D.; Liu, B.; Gao, P.; Wei, W. Andrographolide prevents EV-D68 replication by inhibiting the acidification of virus-containing endocytic vesicles. Front. Microbiol. 2018, 9, 2407. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Xue, H.J.; Ye, W.C.; Fang, B.H.; Liu, Y.H.; Yuan, S.H.; Yu, P.; Wang, Y.Q. Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol. Pharm. Bull. 2009, 32, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ma, Y.B.; Huang, X.Y.; Geng, C.A.; Zhao, Y.; Wang, L.J.; Guo, R.H.; Liang, W.J.; Zhang, X.M.; Chen, J.J. Synthesis, structure-activity relationships and biological evaluation of dehydroandrographolide and andrographolide derivatives as novel anti-hepatitis B virus agents. Bioorganic Med. Chem. Lett. 2014, 24, 2353–2359. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Tseng, C.K.; Young, K.C.; Sun, H.Y.; Wang, S.W.; Chen, W.C.; Lin, C.K.; Wu, Y.H. Andrographolide exerts anti-hepatitis C virus activity by up-regulating haeme oxygenase-1 via the p38 MAPK/Nrf2 pathway in human hepatoma cells. Br. J. Pharmacol. 2014, 171, 237–252. [Google Scholar] [CrossRef]
- Seubsasana, S.; Pientong, C.; Ekalaksananan, T.; Thongchai, S.; Aromdee, C. A Potential andrographolide analogue against the replication of herpes simplex virus type 1 in vero cells. Med. Chem. 2011, 7, 237–244. [Google Scholar] [CrossRef]
- Reddy, V.L.N.; Reddy, S.M.; Ravikanth, V.; Krishnaiah, P.; Goud, T.V.; Rao, T.P.; Ram, T.S.; Gonnade, R.G.; Bhadbhade, M.; Venkateswarlu, Y. A new bis-andrographolide ether from Andrographis paniculata nees and evaluation of anti-HIV activity. Nat. Prod. Res. 2005, 19, 223–230. [Google Scholar] [CrossRef]
- Wintachai, P.; Kaur, P.; Lee, R.C.H.; Ramphan, S.; Kuadkitkan, A.; Wikan, N.; Ubol, S.; Roytrakul, S.; Chu, J.J.H.; Smith, D.R. Activity of andrographolide against chikungunya virus infection. Sci. Rep. 2015, 5, 14179. [Google Scholar] [CrossRef]
- Sa-Ngiamsuntorn, K.; Suksatu, A.; Pewkliang, Y.; Thongsri, P.; Kanjanasirirat, P.; Manopwisedjaroen, S.; Charoensutthivarakul, S.; Wongtrakoongate, P.; Pitiporn, S.; Chaopreecha, J.; et al. Anti-SARS-CoV-2 activity of Andrographis paniculata extract and its major component andrographolide in human lung epithelial cells and cytotoxicity evaluation in major organ cell representatives. J. Nat. Prod. 2021, 84, 1261–1270. [Google Scholar] [CrossRef]
- Semkum, P.; Kaewborisuth, C.; Thangthamniyom, N.; Theerawatanasirikul, S.; Lekcharoensuk, C.; Hansoongnern, P.; Ramasoota, P.; Lekcharoensuk, P. A novel plasmid DNA-based foot and mouth disease virus minigenome for intracytoplasmic mRNA production. Viruses 2021, 13, 1047. [Google Scholar] [CrossRef] [PubMed]
- Theerawatanasirikul, S.; Kuo, C.J.; Phetcharat, N.; Lekcharoensuk, P. In silico and in vitro analysis of small molecules and natural compounds targeting the 3CL protease of feline infectious peritonitis virus. Antivir. Res. 2020, 174, 104697. [Google Scholar] [CrossRef] [PubMed]
- Theerawatanasirikul, S.; Thangthamniyom, N.; Kuo, C.J.; Semkum, P.; Phecharat, N.; Chankeeree, P.; Lekcharoensuk, P. Natural phytochemicals, luteolin and isoginkgetin, inhibit 3C protease and infection of FMDV, in silico and in vitro. Viruses 2021, 13, 2118. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, P.; Wiriyarat, W.; Petcharat, N.; Lekcharoensuk, C.; Auewarakul, P.; Richt, J.A. Cloned cDNA of A/swine/Iowa/15/1930 internal genes as a candidate backbone for reverse genetics vaccine against influenza A viruses. Vaccine 2012, 30, 1453–1459. [Google Scholar] [CrossRef][Green Version]
- Sariya, L.; Thangthumniyom, N.; Wajjwalku, W.; Chumsing, W.; Ramasoota, P.; Lekcharoensuk, P. Expression of foot and mouth disease virus nonstructural polyprotein 3ABC with inactive 3Cpro in Escherichia coli. Protein Expr. Purif. 2011, 80, 17–21. [Google Scholar] [CrossRef]
- Srisombundit, V.; Tungthumniyom, N.; Linchongsubongkoch, W.; Lekcharoensuk, C.; Sariya, L.; Ramasoota, P.; Lekcharoensuk, P. Development of an inactivated 3Cpro-3ABC (mu3ABC) ELISA to differentiate cattle infected with foot and mouth disease virus from vaccinated cattle. J. Virol. Methods 2013, 188, 161–167. [Google Scholar] [CrossRef]
- Du, Y.; Bi, J.; Liu, J.; Liu, X.; Wu, X.; Jiang, P.; Yoo, D.; Zhang, Y.; Wu, J.; Wan, R.; et al. 3Cpro of Foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 Nuclear Translocation. J. Virol. 2014, 88, 4908–4920. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Theerawatanasirikul, S.; Lekcharoensuk, P. Virtual Screening of Natural Compounds Targeting Proteases of Coronaviruses and Picornaviruses. In Methods in Pharmacology and Toxicology; Roy, K., Ed.; Springer: New York, NY, USA, 2021; pp. 661–681. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo–distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Panraksa, P.; Ramphan, S.; Khongwichit, S.; Smith, D.R. Activity of andrographolide against dengue virus. Antivir. Res. 2017, 139, 69–78. [Google Scholar] [CrossRef]
- Wiart, C.; Kumar, K.; Yusof, M.Y.; Hamimah, H.; Fauzi, Z.M.; Sulaiman, M. Antiviral properties of ent-labdene diterpenes of Andrographis paniculata Nees, inhibitors of herpes simplex virus type 1. Phytother. Res. 2005, 19, 1069–1070. [Google Scholar] [CrossRef]
- Shi, T.H.; Huang, Y.L.; Chen, C.C.; Pi, W.C.; Hsu, Y.L.; Lo, L.C.; Chen, W.Y.; Fu, S.L.; Lin, C.H. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem. Biophys. Res. Commun. 2020, 533, 467–473. [Google Scholar] [CrossRef]
- Roqué Rosell, N.R.; Mokhlesi, L.; Milton, N.E.; Sweeney, T.R.; Zunszain, P.A.; Curry, S.; Leatherbarrow, R.J. Design and synthesis of irreversible inhibitors of foot-and-mouth disease virus 3C protease. Bioorganic Med. Chem. Lett. 2014, 24, 490–494. [Google Scholar] [CrossRef][Green Version]
- Zunszain, P.A.; Knox, S.R.; Sweeney, T.R.; Yang, J.; Roqué-Rosell, N.; Belsham, G.J.; Leatherbarrow, R.J.; Curry, S. Insights into cleavage specificity from the crystal structure of foot-and-mouth disease virus 3C protease complexed with a peptide substrate. J. Mol. Biol. 2010, 395, 375–389. [Google Scholar] [CrossRef]
- Li, F.; Lee, E.M.; Sun, X.; Wang, D.; Tang, H.; Zhou, G.C. Design, synthesis and discovery of andrographolide derivatives against Zika virus infection. Eur. J. Med. Chem. 2020, 187, 111925. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Ohashi, S.; Reiko, Y.; Kazumi, T.; Furuta, Y. The inhibition of FMD virus excretion from the infected pigs by an antiviral agent, T-1105. In FAO Report of the Research Group of the Standing Technical Committee of European Commission for the control of Foot-and-Mouth Disease; Appendix 64; FAO: Paphos, Cyprus, 2006; pp. 418–424. [Google Scholar]
- Hayden, F.G.; Turner, R.B.; Gwaltney, J.M.; Chi-Burris, K.; Gersten, M.; Hsyu, P.; Patick, A.K.; Smith, G.J.; Zalman, L.S. Phase II, Randomized, Double-Blind, Placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003, 47, 3907–3916. [Google Scholar] [CrossRef] [PubMed]
- Binford, S.L.; Weady, P.T.; Maldonado, F.; Brothers, M.A.; Matthews, D.A.; Patick, A.K. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2007, 51, 4366–4373. [Google Scholar] [CrossRef] [PubMed]
- Chungsamarnyart, N.; Sirinarumitr, T.; Chumsing, W.; Wajjawalku, W. In vitro study of antiviral activity of plant crude-extracts against the foot and mouth disease virus. Kasetsart J. (Nat. Sci.) 2007, 41, 97–103. [Google Scholar]
- Abou Assi, R.; Darwis, Y.; Abdulbaqi, I.M.; Khan, A.A.; Vuanghao, L.; Laghari, M.H. Morinda citrifolia (Noni): A comprehensive review on its industrial uses, pharmacological activities, and clinical trials. Arab. J. Chem. 2017, 10, 691–707. [Google Scholar] [CrossRef]
- Lalani, S.; Poh, C.L. Flavonoids as antiviral agents for enterovirus A71 (EV-A71). Viruses 2020, 12, 184. [Google Scholar] [CrossRef]
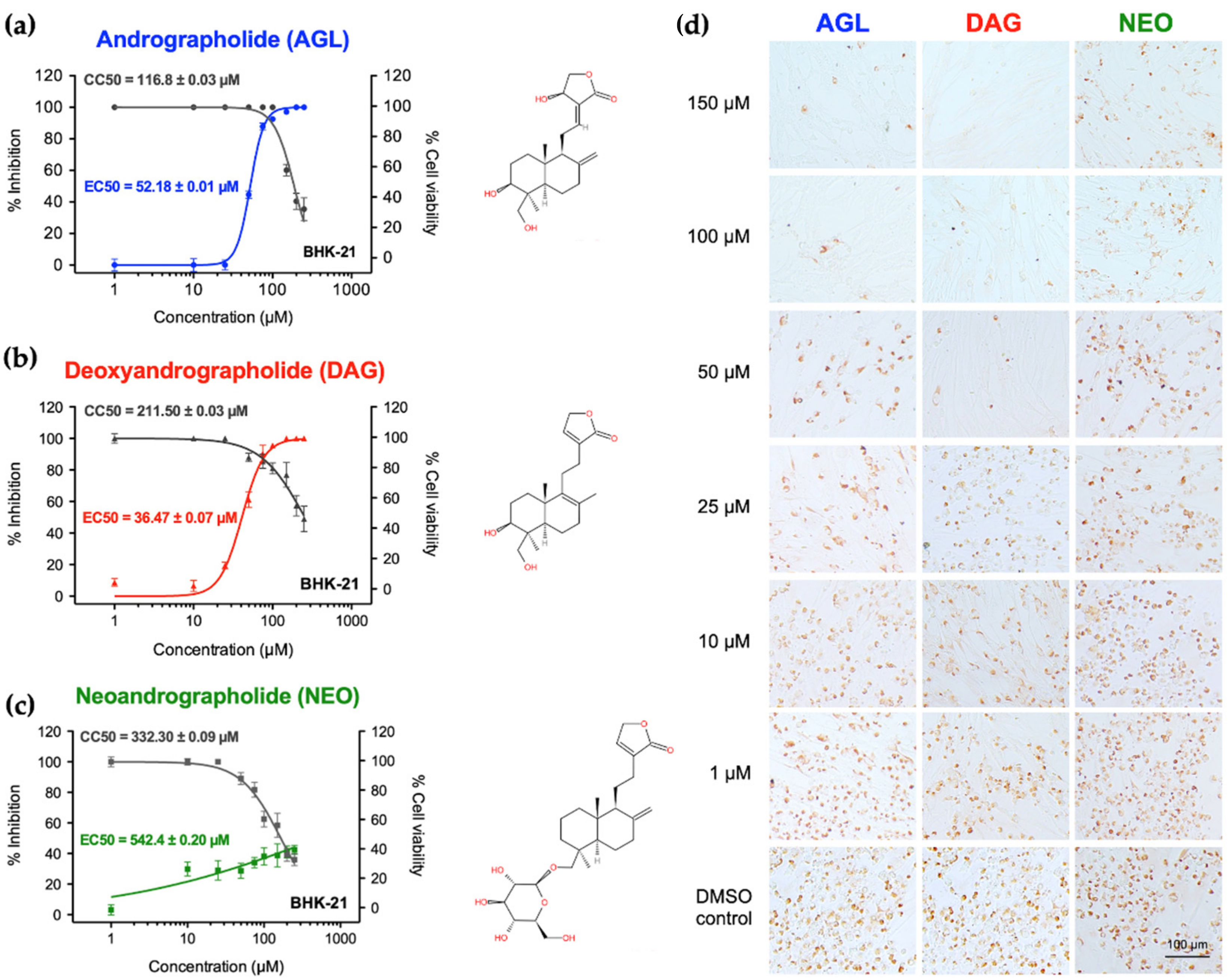
| Cytotoxicity (µM) | AGL | DAG | NEO | |
|---|---|---|---|---|
| BHK-21 | CC50 1 | 116.8 ± 0.03 | 332.3 ± 0.09 | 211.5 ± 0.03 |
| CC10 2 | 94.55 ± 0.02 | 81.05 ± 0.01 | 149.94 ± 0.21 | |
| HEK 293T | CC50 1 | 453.00 ± 0.04 | 651.40 ± 0.32 | 914.20 ± 0.04 |
| CC10 2 | 259.00 ± 0.11 | 155.55 ± 0.91 | 270.60 ± 0.03 | |
| Andrographolides | IPMA | Intracellular Protease Assay | ||
|---|---|---|---|---|
| EC50 (µM) 1 | SI 2 | IC50 (µM) 3 | IC90 (µM) 4 | |
| AGL | 52.18 ± 0.01 | 2.23 | 67.43± 0.81 | 267.21 ± 2.43 |
| (CC10 5; 259.00 ± 0.11) | ||||
| DAG | 36.47 ± 0.07 | 9.11 | 25.58 ± 1.41 | 122.88 ± 2.09 |
| (CC10; 155.55 ± 0.91) | ||||
| NEO | 542.40 ± 0.20 | 0.38 | 111.4 ± 2.04 | 635.76 ± 2.90 |
| (CC10; 270 ± 0.03) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theerawatanasirikul, S.; Lueangaramkul, V.; Thangthamniyom, N.; Chankeeree, P.; Semkum, P.; Lekcharoensuk, P. Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3Cpro. Animals 2022, 12, 1995. https://doi.org/10.3390/ani12151995
Theerawatanasirikul S, Lueangaramkul V, Thangthamniyom N, Chankeeree P, Semkum P, Lekcharoensuk P. Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3Cpro. Animals. 2022; 12(15):1995. https://doi.org/10.3390/ani12151995
Chicago/Turabian StyleTheerawatanasirikul, Sirin, Varanya Lueangaramkul, Nattarat Thangthamniyom, Penpitcha Chankeeree, Ploypailin Semkum, and Porntippa Lekcharoensuk. 2022. "Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3Cpro" Animals 12, no. 15: 1995. https://doi.org/10.3390/ani12151995
APA StyleTheerawatanasirikul, S., Lueangaramkul, V., Thangthamniyom, N., Chankeeree, P., Semkum, P., & Lekcharoensuk, P. (2022). Andrographolide and Deoxyandrographolide Inhibit Protease and IFN-Antagonist Activities of Foot-and-Mouth Disease Virus 3Cpro. Animals, 12(15), 1995. https://doi.org/10.3390/ani12151995






