Transcriptomics and Metabolomics Analysis of the Ovaries of High and Low Egg Production Chickens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animal and Sample Collection
2.3. Metabolite Extraction, Detection, and Analysis
2.4. RNA Sequencing (RNA-Seq) and Data Analysis
2.5. GO and KEGG Enrichment Analysis of DEGs
2.6. Statistical Analysis
3. Results
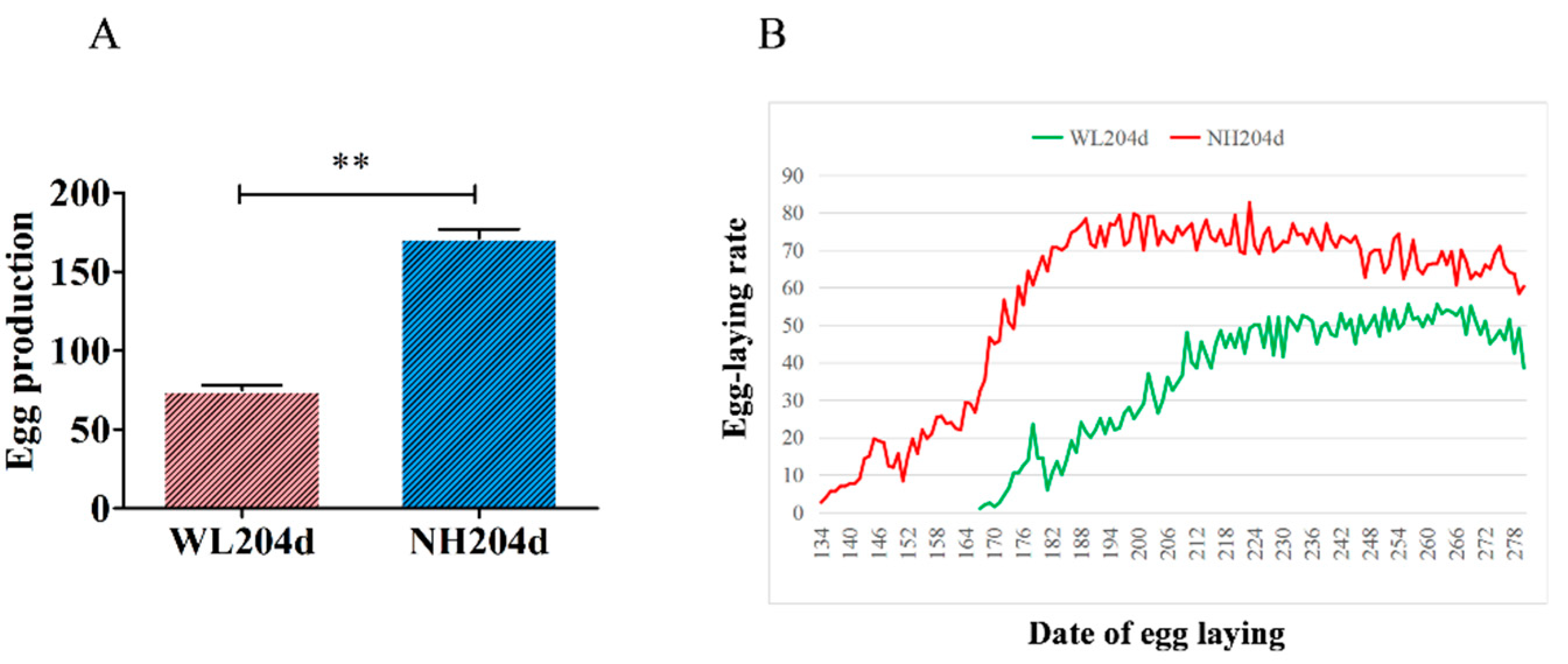
3.1. Production Performance
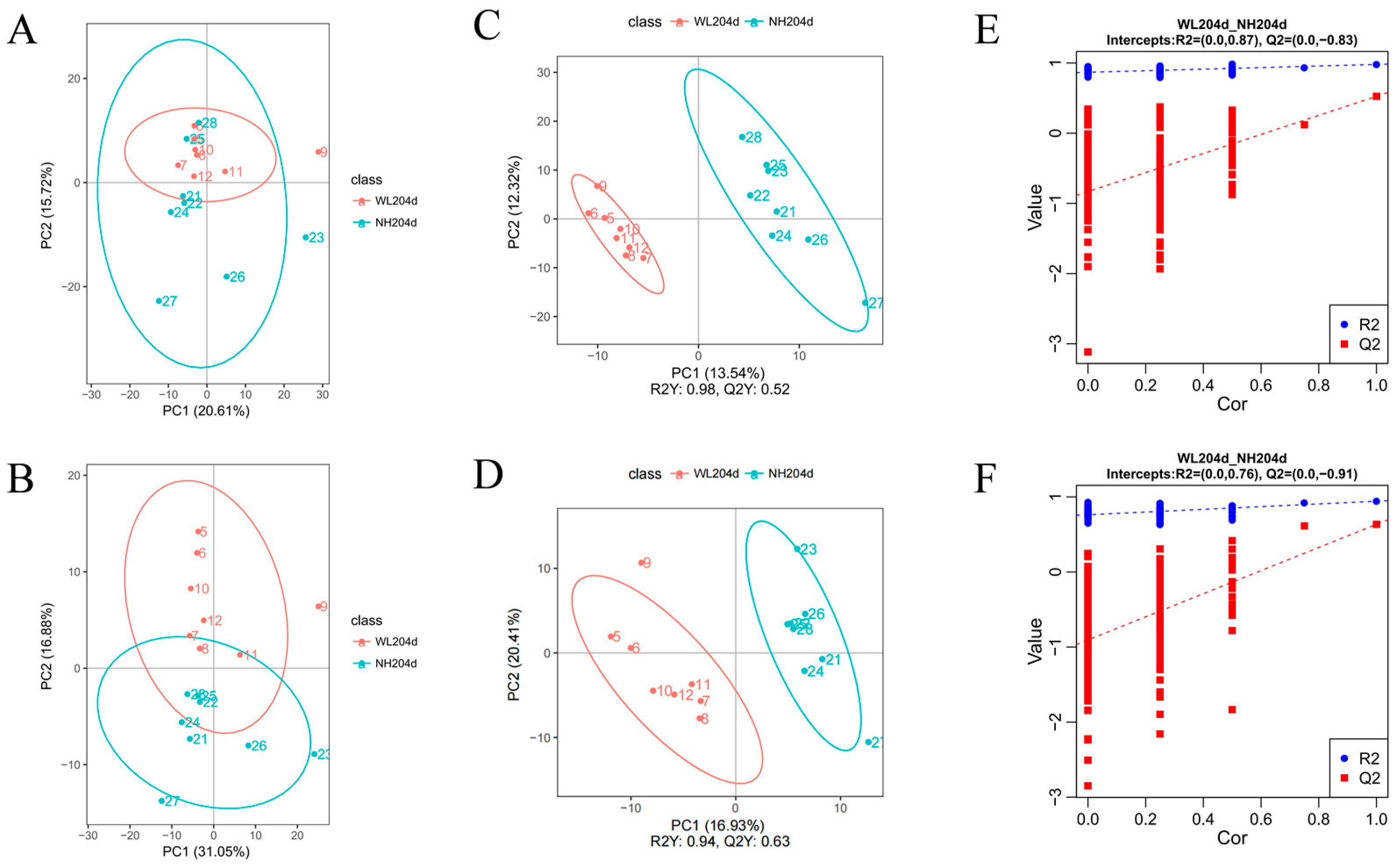
3.2. Quality Control, Principal Component Analyses of Metabolomics
3.3. Metabolomics Analysis of High- and Low-Production Groups
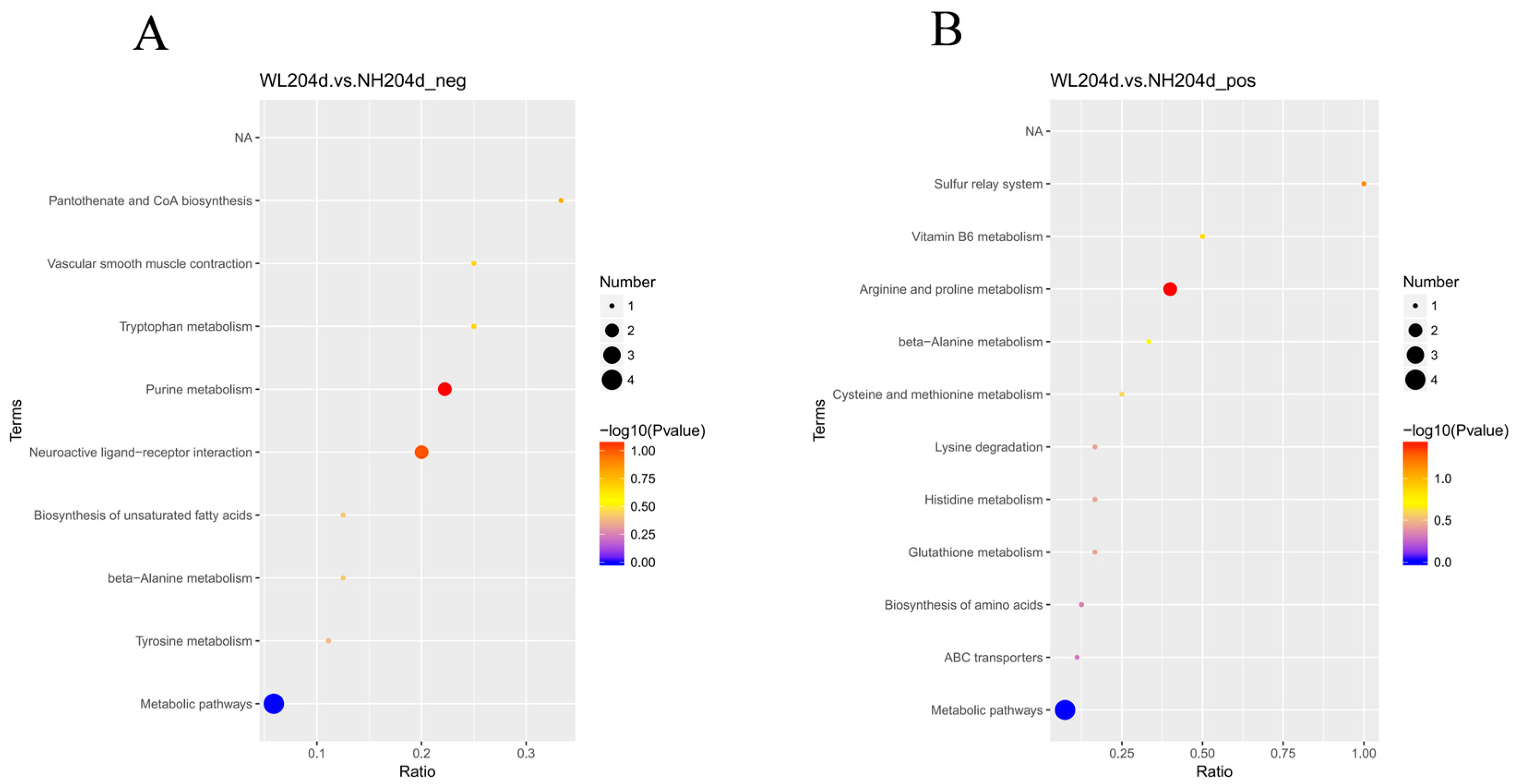
3.4. KEGG Analysis of SDMs
3.5. Transcriptomics Analysis of High- and Low-Production Groups
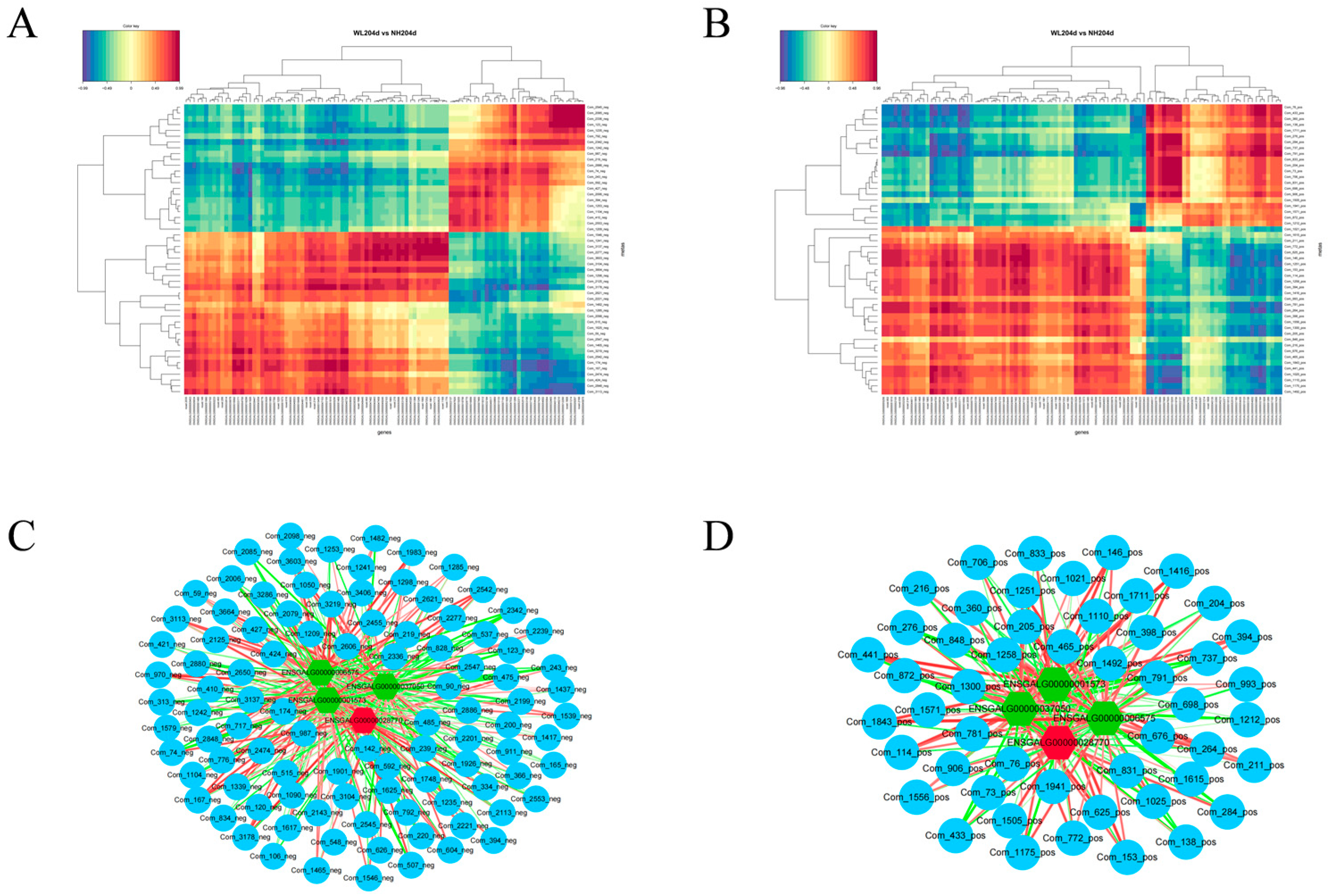
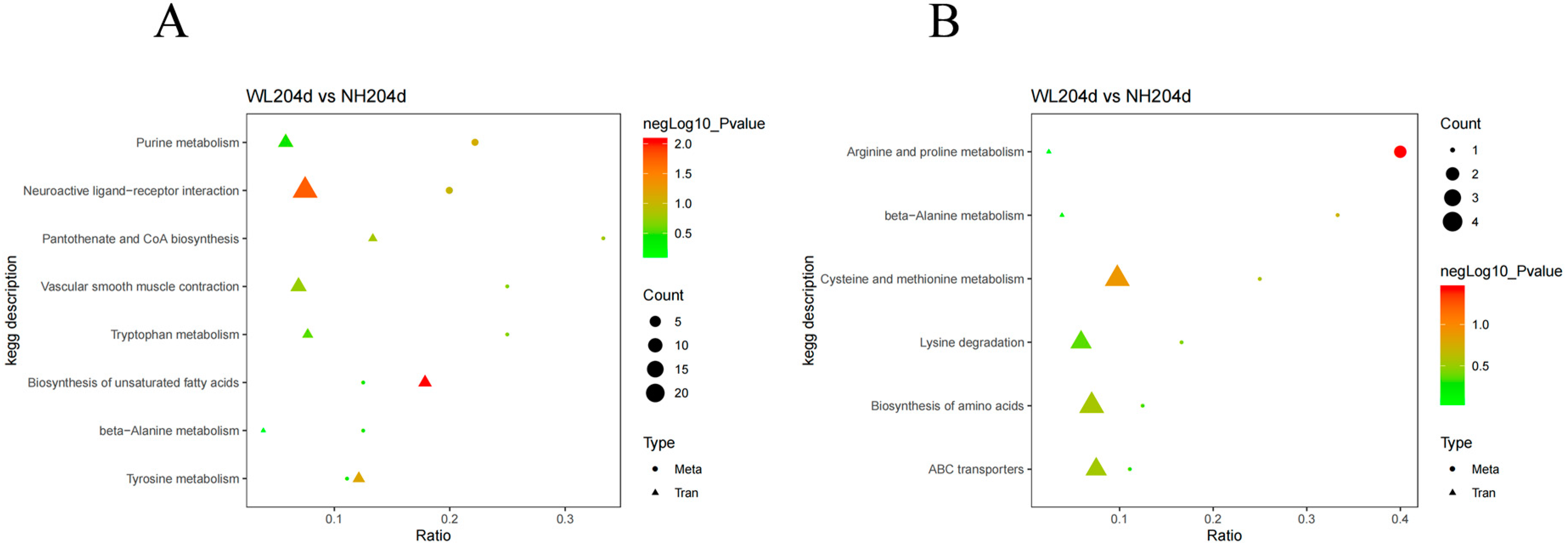
3.6. Comprehensive Analysis of the Transcriptomics and Metabolomics
4. Discussion
4.1. Transcriptomics Analysis
4.2. Metabolomics Analysis
4.3. Combining Analysis of Transcriptomics and Metabolomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woodruff, T.K.; Shea, L.D. The role of the extracellular matrix in ovarian follicle development. Reprod. Sci. 2007, 14, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Biscarini, F.; Bovenhuis, H.; Ellen, E.D.; Addo, S.; van Arendonk, J.A. Estimation of heritability and breeding values for early egg production in laying hens from pooled data. Poult. Sci. 2010, 89, 1842–1849. [Google Scholar] [CrossRef]
- Sultan, M.; Schulz, M.H.; Richard, H.; Magen, A.; Klingenhoff, A.; Scherf, M.; Seifert, M.; Borodina, T.; Soldatov, A.; Parkhomchuk, D.; et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008, 321, 956–960. [Google Scholar] [CrossRef] [Green Version]
- Bao, X.Y.; Song, Y.P.; Li, T.; Zhang, S.S.; Huang, L.H.; Zhang, S.Y.; Cao, J.T.; Liu, X.L.; Zhang, J.Q. Comparative Transcriptome Profiling of Ovary Tissue between Black Muscovy Duck and White Muscovy Duck with High- and Low-Egg Production. Genes 2021, 12, 57. [Google Scholar] [CrossRef]
- Yang, G.; Li, S.M.; Zhao, Q.Q.; Chu, J.Y.; Zhou, B.G.; Fan, S.J.; Shi, F.Y.; Wei, X.R.; Hu, X.W.; Zheng, X.T.; et al. Transcriptomic and metabolomic insights into the variety of sperm storage in oviduct of egg layers. Poultry Sci. 2021, 100, 101087. [Google Scholar] [CrossRef]
- Wu, X.; Pan, X.L.; Cao, S.M.; Xu, F.Q.; Lan, L.M.; Zhang, Y.Y.; Lian, S.Y.; Yan, M.J.; Li, A. iTRAQ-based quantitative proteomic analysis provides insights into strong broodiness in Muscovy duck (Cairina moschata) combined with metabolomics analysis. J. Proteom. 2019, 204, 103401. [Google Scholar] [CrossRef]
- Yuan, X.; Hu, S.; Li, L.; Liu, H.; He, H.; Wang, J. Metabolomic Analysis of SCD during Goose Follicular Development: Implications for Lipid Metabolism. Genes 2020, 11, 1001. [Google Scholar] [CrossRef]
- Zhan, H.; Xiong, Y.; Wang, Z.; Dong, W.; Zhou, Q.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Integrative analysis of transcriptomic and metabolomic profiles reveal the complex molecular regulatory network of meat quality in Enshi black pigs. Meat Sci. 2022, 183, 108642. [Google Scholar] [CrossRef]
- Sun, H.Z.; Wang, D.M.; Liu, H.Y.; Liu, J.X. Metabolomics Integrated with Transcriptomics Reveals a Subtle Liver Metabolic Risk in Dairy Cows Fed Different Crop By-products. Proteomics 2018, 18, e1800122. [Google Scholar] [CrossRef]
- Dou, T.; Yan, S.; Liu, L.; Wang, K.; Jian, Z.; Xu, Z.; Zhao, J.; Wang, Q.; Sun, S.; Talpur, M.Z.; et al. Integrative analysis of transcriptomics and metabolomics to reveal the melanogenesis pathway of muscle and related meat characters in Wuliangshan black-boned chickens. BMC Genom. 2022, 23, 173. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach To Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, L.; Han, K.; Zhang, X.; Zhang, G.; Dai, G.; Wang, J.; Xie, K. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Anim. Reprod. Sci. 2019, 208, 106114. [Google Scholar] [CrossRef]
- Tao, Z.; Song, W.; Zhu, C.; Xu, W.; Liu, H.; Zhang, S.; Huifang, L. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 2017, 96, 4378–4388. [Google Scholar] [CrossRef]
- Ouyang, Q.Y.; Hu, S.Q.; Wang, G.S.; Hu, J.W.; Zhang, J.M.; Li, L.; Hu, B.; He, H.; Liu, H.H.; Xia, L.; et al. Comparative Transcriptome Analysis Suggests Key Roles for 5-Hydroxytryptamlne Receptors in Control of Goose Egg Production. Genes 2020, 11, 455. [Google Scholar] [CrossRef] [Green Version]
- Luan, X.H.; Liu, D.W.; Cao, Z.Z.; Luo, L.N.; Liu, M.; Gao, M.; Zhang, X.Y. Transcriptome Profiling Identifies Differentially Expressed Genes in Huoyan Goose Ovaries between the Laying Period and Ceased Period. PLoS ONE 2014, 9, e113211. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.X.; Sun, X.; Chimbaka, I.M.; Qin, N.; Xu, X.X.; Liswaniso, S.; Xu, R.F.; Gonzalez, J.M. Transcriptome Analysis of Ovarian Follicles Reveals Potential Pivotal Genes Associated With Increased and Decreased Rates of Chicken Egg Production. Front. Genet. 2021, 12, 622751. [Google Scholar] [CrossRef]
- Zhang, G.X.; Fan, Q.C.; Wang, J.Y.; Zhang, T.; Xue, Q.; Shi, H.Q. Genome-wide association study on reproductive traits in Jinghai Yellow Chicken. Anim. Reprod. Sci. 2015, 163, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Xiao, C.; Deng, J.X.; Yang, Z.L.; Zou, L.Q.; Du, W.Y.; Li, S.X.; Huo, X.Q.; Zeng, L.H.; Yang, X.R. Transcriptome analysis reveals key genes and pathways associated with egg production in Nandan-Yao domestic chicken. Comp. Biochem. Phys. D 2021, 40, 100889. [Google Scholar] [CrossRef] [PubMed]
- Bello, S.F.; Xu, H.P.; Guo, L.J.; Li, K.; Zheng, M.; Xu, Y.B.; Zhang, S.Y.; Bekele, E.J.; Bahareldin, A.A.; Zhu, W.J.; et al. Hypothalamic and ovarian transcriptome profiling reveals potential candidate genes in low and high egg production of white Muscovy ducks (Cairina moschata). Poult. Sci. 2021, 100, 101310. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Baldwin, J.M.; Roger, S.; Baldwin, S.A. Insights into the molecular mechanisms underlying mammalian P2X7 receptor functions and contributions in diseases, revealed by structural modeling and single nucleotide polymorphisms. Front. Pharmacol. 2013, 4, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, C.W.; Choong, Y.-T.; Short, J.L.; Exintaris, B.; Malone, D.T.; Allen, A.M.; Evans, R.J.; Ventura, S. Male contraception via simultaneous knockout of α1A-adrenoceptors and P2X1-purinoceptors in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 20825–20830. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Maitra, A.; Ahlawat, S.; Roy, M.; Mandakmale, S.; Tantia, M.S. Identification of novel SNPs in INHBB gene of Indian goat. Indian J. Anim. Sci. 2015, 85, 55–59. [Google Scholar]
- M’Baye, M.; Hua, G.H.; Khan, H.A.; Yang, L.G. RNAi-mediated knockdown of INHBB increases apoptosis and inhibits steroidogenesis in mouse granulosa cells. J. Reprod. Develop. 2015, 61, 391–397. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.F.; He, X.Y.; Zhang, X.S.; Zhang, J.L.; Guo, X.F.; Sun, W.; Chu, M.X. Analysis of Expression Profiles of CircRNA and MiRNA in Oviduct during the Follicular and Luteal Phases of Sheep with Two Fecundity (FecB Gene) Genotypes. Animals 2021, 11, 2826. [Google Scholar] [CrossRef]
- Conconi, M.T.; Spinazzi, R.; Nussdorfer, G.G. Endogenous ligands of PACAP/VIP receptors in the autocrine-paracrine regulation of the adrenal gland. Int. Rev. Cytol. 2006, 249, 1–51. [Google Scholar] [CrossRef]
- Sun, X.; Chen, X.X.; Zhao, J.H.; Ma, C.; Yan, C.C.; Liswaniso, S.; Xu, R.F.; Qin, N. Transcriptome comparative analysis of ovarian follicles reveals the key genes and signaling pathways implicated in hen egg production. BMC Genom. 2021, 22, 899. [Google Scholar] [CrossRef]
- Shao, F.; Bao, H.G.; Li, H.W.; Duan, J.L.; Li, J.Y.; Ling, Y.; Wu, C.X. Ovary removal modifies liver message RNA profiles in single Comb White Leghorn chickens. Poult. Sci. 2020, 99, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Feng, T.; DeVore, A.A.; Perego, M.C.; Morrell, B.C.; Spicer, L.J. Effects of N-carbamylglutamate and arginine on steroidogenesis and proliferation of pig granulosa cells in vitro. Anim. Reprod. Sci. 2019, 209, 106138. [Google Scholar] [CrossRef]
- Luderer, U.; Kavanagh, T.J.; White, C.C.; Faustman, E.M. Gonadotropin regulation of glutathione synthesis in the rat ovary. Reprod. Toxicol. 2001, 15, 495–504. [Google Scholar] [CrossRef]
- Kim, Y.; Han, D.; Min, H.; Jin, J.; Yi, E.C.; Kim, Y. Comparative Proteomic Profiling of Pancreatic Ductal Adenocarcinoma Cell Lines. Mol. Cells 2014, 37, 888–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manuck, T.A. 17-alpha hydroxyprogesterone caproate for preterm birth prevention: Where have we been, how did we get here, and where are we going? Semin. Perinatol. 2017, 41, 461–467. [Google Scholar] [CrossRef]
- Kowalczyk-Zieba, I.; Boruszewska, D.; Suwik, K.; Staszkiewicz-Chodor, J.; Jaworska, J.; Woclawek-Potocka, I. Iloprost affects in vitro maturation and developmental competence of bovine oocytes. Theriogenology 2020, 157, 286–296. [Google Scholar] [CrossRef]
- Kim, J.S.; Chae, J.I.; Song, B.S.; Lee, K.S.; Choo, Y.K.; Chang, K.T.; Park, H.; Koo, D.B. Iloprost, a prostacyclin analogue, stimulates meiotic maturation and early embryonic development in pigs. Reprod. Fertil. Dev. 2010, 22, 437–447. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. The functional role of polyamines in eukaryotic cells. Int. J. Biochem. Cell. Biol. 2019, 107, 104–115. [Google Scholar] [CrossRef]
- Fernandes, J.R.D.; Jain, S.; Banerjee, A. Expression of ODC1, SPD, SPM and AZIN1 in the hypothalamus, ovary and uterus during rat estrous cycle. Gen. Comp. Endocrinol. 2017, 246, 9–22. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Chi, Y.J.; Yu, Y.S.; Liu, J.L.; Su, R.W.; Ma, X.H.; Shan, C.H.; Yang, Z.M. Polyamines are essential in embryo implantation: Expression and function of polyamine-related genes in mouse uterus during peri-implantation period. Endocrinology 2008, 149, 2325–2332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Sallaberry, C.; Simmen, F.A.; Simmen, R.C. Polyamine- and insulin-like growth factor-I-mediated proliferation of porcine uterine endometrial cells: A potential role for spermidine/spermine N(1)-acetyltransferase during peri-implantation. Biol. Reprod. 2001, 65, 587–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheth, S.; Brito, R.; Mukherjea, D.; Rybak, L.P.; Ramkumar, V. Adenosine receptors: Expression, function and regulation. Int. J. Mol. Sci. 2014, 15, 2024–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Sample | Raw_Reads | Clean_Reads | Total-Map | Error_Rate | Q30 | GC_Pct |
|---|---|---|---|---|---|---|
| WL204d_1 | 59,739,580 | 58,953,024 | 54,412,280 (92.3%) | 0.02% | 94.49% | 51.2% |
| WL204d_2 | 58,473,150 | 57,710,008 | 52,207,089 (90.46%) | 0.02% | 94.4% | 51.23% |
| WL204d_3 | 61,333,900 | 60,429,438 | 54,577,702 (90.32%) | 0.03% | 94.07% | 51.33% |
| WL204d_4 | 57,835,138 | 56,950,072 | 51,440,969 (90.33%) | 0.03% | 94.2% | 50.71% |
| NH204d_1 | 59,434,470 | 58,593,266 | 53,253,424 (90.89%) | 0.03% | 94.22% | 50.91% |
| NH204d_2 | 53,435,540 | 52,503,528 | 47,500,358 (90.47%) | 0.03% | 94.22% | 50.53% |
| NH204d_3 | 62,255,842 | 61,476,260 | 56,020,220 (91.12%) | 0.03% | 94.17% | 51.38% |
| NH204d_4 | 53,250,580 | 52,498,894 | 46,560,294 (88.69%) | 0.03% | 94.17% | 51.69% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Zhang, H.; Cao, H.; Zhou, W.; Xiang, X.; Yin, Z. Transcriptomics and Metabolomics Analysis of the Ovaries of High and Low Egg Production Chickens. Animals 2022, 12, 2010. https://doi.org/10.3390/ani12162010
Huang X, Zhang H, Cao H, Zhou W, Xiang X, Yin Z. Transcriptomics and Metabolomics Analysis of the Ovaries of High and Low Egg Production Chickens. Animals. 2022; 12(16):2010. https://doi.org/10.3390/ani12162010
Chicago/Turabian StyleHuang, Xuan, Haiyang Zhang, Haiyue Cao, Wei Zhou, Xin Xiang, and Zhaozheng Yin. 2022. "Transcriptomics and Metabolomics Analysis of the Ovaries of High and Low Egg Production Chickens" Animals 12, no. 16: 2010. https://doi.org/10.3390/ani12162010
APA StyleHuang, X., Zhang, H., Cao, H., Zhou, W., Xiang, X., & Yin, Z. (2022). Transcriptomics and Metabolomics Analysis of the Ovaries of High and Low Egg Production Chickens. Animals, 12(16), 2010. https://doi.org/10.3390/ani12162010






