Expression of Key Factors of the Hippo Signaling Pathway in Yak (Bos grunniens) Mammary Gland
Abstract
:Simple Summary
Abstract
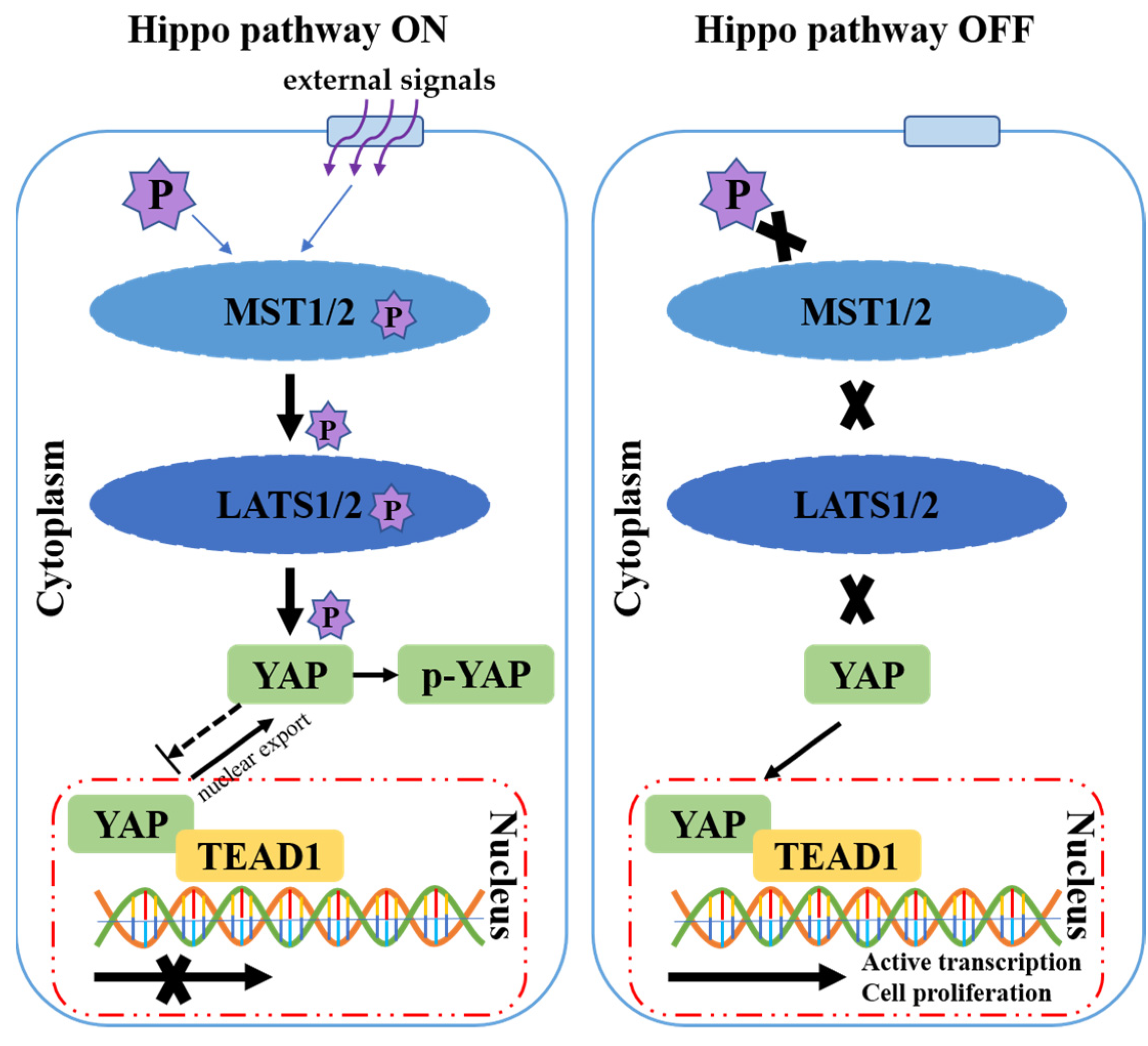
1. Introduction
2. Materials and Methods
2.1. Collection of Yak Mammary Gland Tissue Samples
2.2. Extraction of Total RNA and qRT-PCR Detection
2.3. Total Protein Was Extracted and Detected by WB
2.4. Preparation of Paraffin Sections and Immunohistochemical Localization
2.5. Data Analysis
3. Results
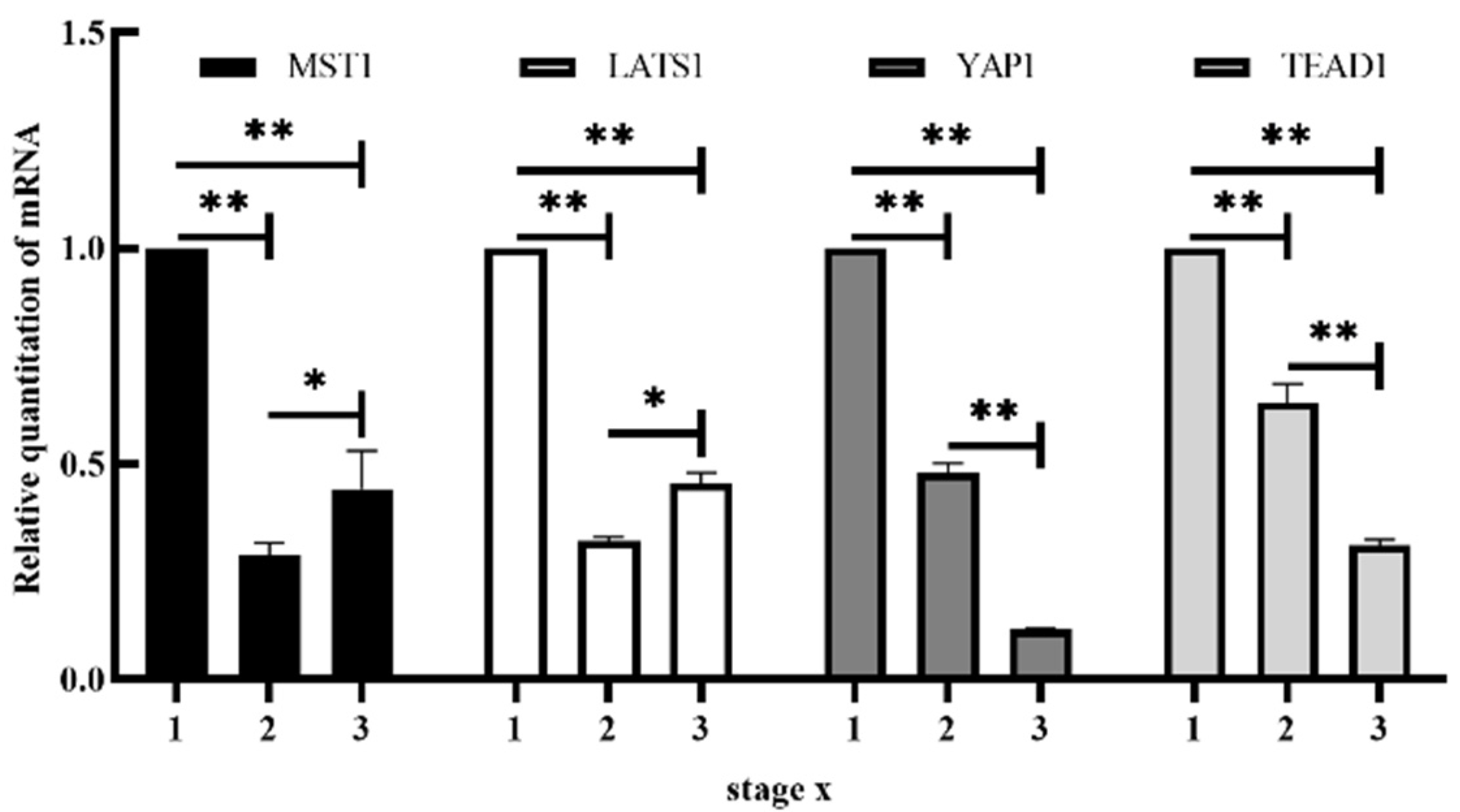
3.1. mRNA Expression of MST1, LATS1, YAP1 and TEAD1
3.2. Expression of MST1, LATS1, YAP1 and TEAD1 Proteins
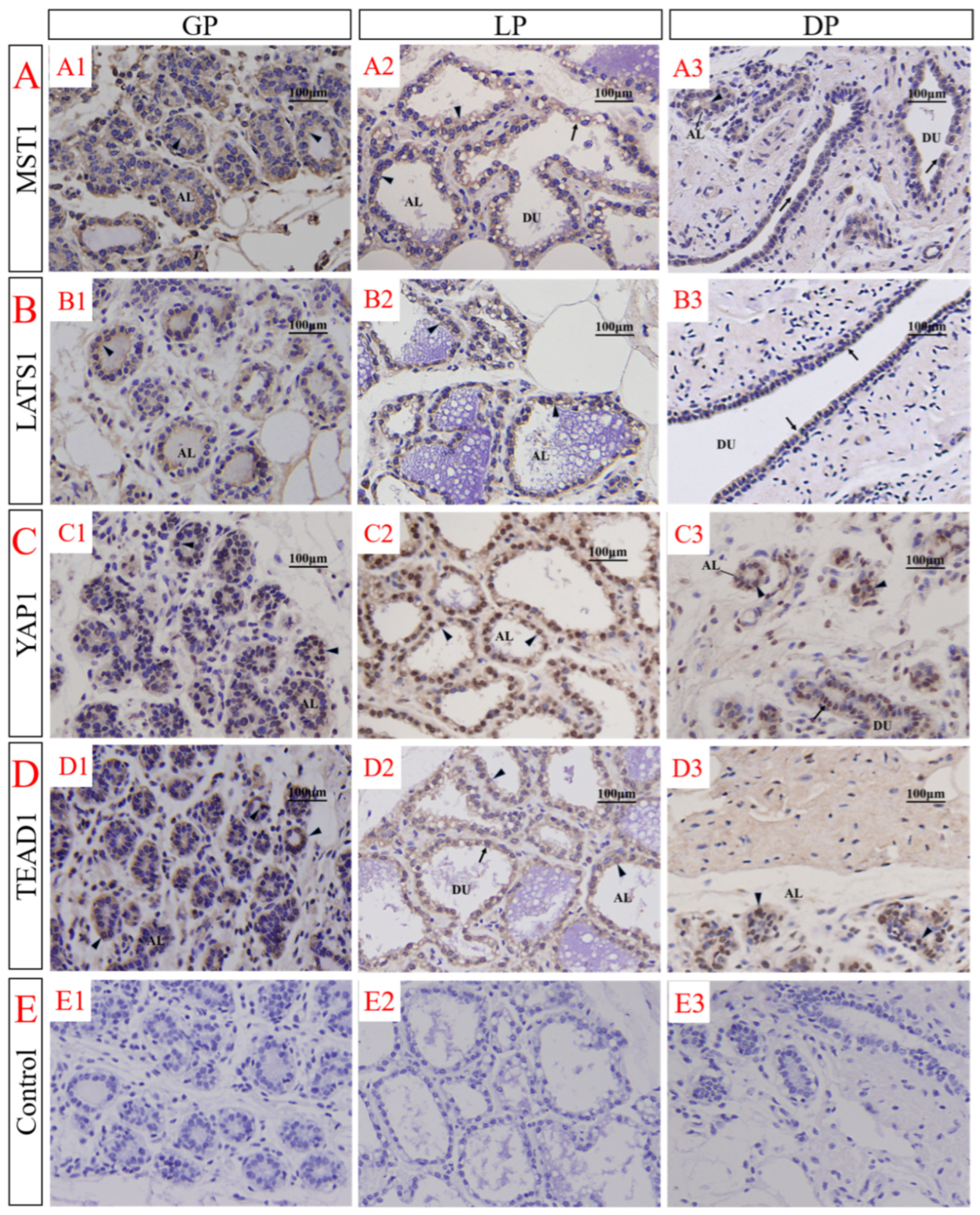
3.3. Immunolocalization of MST1, LATS1, YAP1 and TEAD1 in the Female Yak’s Mammary Gland
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pantalacci, S.; Tapon, N.; Léopold, P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003, 5, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Pfleger, C.M.; Hariharan, I.K. The drosophila mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 2003, 114, 457–467. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. Hippo encodes a ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, W.; Wang, B.; Trinko, R.; Jiang, J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003, 17, 2514–2519. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.C.; Wei, X.; Shimizu, T.; Ramos, E.; Rohrbaugh, M.; Nikolaidis, N.; Ho, L.L.; Li, Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 2005, 120, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, S.; Barrera, J.; Matthews, K.; Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005, 122, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mak, K.K.; Topol, L.; Yun, K.; Hu, J.; Garrett, L.; Chen, Y.; Park, O.; Chang, J.; Simpson, R.M.; et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA 2010, 107, 1431–1436. [Google Scholar] [CrossRef]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2008, 17, 2054–2060. [Google Scholar] [CrossRef] [PubMed]
- Barry, E.R.; Camargo, F.D. The Hippo superhighway: Signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr. Opin. Cell Biol. 2013, 25, 247–253. [Google Scholar] [CrossRef]
- Gumbiner, B.M.; Kim, N.G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 2014, 127, 709–717. [Google Scholar] [CrossRef]
- Halder, G.; Johnson, R.L. Hippo signaling: Growth control and beyond. Development 2011, 138, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Harvey, K.F.; Hariharan, I.K. The hippo pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011288. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Lai, Z.C. Mutual regulation between Hippo signaling and actin cytoskeleton. Protein Cell 2013, 4, 904–910. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef]
- Staley, B.K.; Irvine, K.D. Hippo signaling in Drosophila: Recent advances and insights. Dev. Dyn. 2012, 241, 3–15. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef]
- Lin, C.; Yao, E.; Chuang, P.T. A conserved MST1/2–YAP axis mediates Hippo signaling during lung growth. Dev. Biol. 2015, 403, 101–113. [Google Scholar] [CrossRef]
- Chan, E.H.; Nousiainen, M.; Chalamalasetty, R.B.; Schäfer, A.; Nigg, E.A.; Silljé, H.H. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 2005, 24, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Mazack, V.; Sudol, M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J. Biol. Chem. 2008, 283, 27534–27546. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Tumaneng, K.; Wang, C.Y.; Guan, K.L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 2010, 24, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Jacquemin, P.; Hwang, J.J.; Martial, J.A.; Dollé, P.; Davidson, I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J. Biol. Chem. 1996, 271, 21775–21785. [Google Scholar] [CrossRef]
- Ota, M.; Sasaki, H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 2008, 135, 4059–4069. [Google Scholar] [CrossRef]
- Sawada, A.; Kiyonari, H.; Ukita, K.; Nishioka, N.; Imuta, Y.; Sasaki, H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol. Cell Biol. 2008, 28, 3177–3189. [Google Scholar] [CrossRef]
- Reginensi, A.; Scott, R.P.; Gregorieff, A.; Bagherie-Lachidan, M.; Chung, C.; Lim, D.S.; Pawson, T.; Wrana, J.; McNeill, H. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013, 9, e1003380. [Google Scholar] [CrossRef]
- Schlegelmilch, K.; Mohseni, M.; Kirak, O.; Pruszak, J.; Rodriguez, J.R.; Zhou, D.; Kreger, B.T.; Vasioukhin, V.; Avruch, J.; Brummelkamp, T.R.; et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 2011, 144, 782–795. [Google Scholar] [CrossRef]
- Xin, M.; Kim, Y.; Sutherland, L.B.; Qi, X.; McAnally, J.; Schwartz, R.J.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 2011, 4, ra70. [Google Scholar] [CrossRef]
- Zhang, N.; Bai, H.; David, K.K.; Dong, J.; Zheng, Y.; Cai, J.; Giovannini, M.; Liu, P.; Anders, R.A.; Pan, D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 2010, 19, 27–38. [Google Scholar] [CrossRef]
- Wang, X.; Su, L.; Ou, Q. Yes-associated protein promotes tumour development in luminal epithelial derived breast cancer. Eur. J. Cancer 2012, 48, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Pepling, M.E.; Diaz, F.J. Lats1 Deletion Causes Increased Germ Cell Apoptosis and Follicular Cysts in Mouse Ovaries. Biol. Reprod. 2015, 22, 1–11. [Google Scholar] [CrossRef]
- Yuan, M.; Tomlinson, V.; Lara, R.; Holliday, D.; Chelala, C.; Harada, T.; Gangeswaran, R.; Manson-Bishop, C.; Smith, P.; Danovi, S.A.; et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008, 15, 1752–1759. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, N.; Gray, R.S.; Li, H.; Ewald, A.J.; Zahnow, C.A.; Pan, D. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 2014, 28, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Lamar, J.M.; Stern, P.; Liu, H.; Schindler, J.W.; Jiang, Z.G.; Hynes, R.O. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. USA 2012, 109, E2441–E2450. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Lei, Q.; Guan, K.L. The Hippo–YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 2010, 24, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chang, T.; Wang, Y.; Liu, Y.; Li, W.; Li, M.; Fan, H.Y. YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PLoS ONE 2014, 9, e91770. [Google Scholar] [CrossRef]
- Jiangfeng, F.; Yuzhu, L.; Sijiu, Y.; Yan, C.; Gengquan, X.; Libin, W.; Yangyang, P.; Honghong, H. Transcriptional profiling of two different physiological states of the yak mammary gland using RNA sequencing. PLoS ONE 2018, 13, e0201628. [Google Scholar] [CrossRef]
- Fan, J.; Yu, Y.; Han, X.; He, H.; Luo, Y.; Yu, S.; Cui, Y.; Xu, G.; Wang, L.; Pan, Y. The expression of hypoxia-inducible factor-1 alpha in primary reproductive organs of the female yak (Bos grunniens) at different reproductive stages. Reprod. Domest. Anim. 2020, 55, 1371–1382. [Google Scholar] [CrossRef]
- Ding, L.; Wang, Y.; Kreuzer, M.; Guo, X.; Mi, J.; Gou, Y.; Shang, Z.; Zhang, Y.; Zhou, J.; Wang, H.; et al. Seasonal variations in the fatty acid profile of milk from yaks grazing on the Qinghai-Tibetan plateau. J. Dairy Res. 2013, 80, 410–417. [Google Scholar] [CrossRef]
- LI, Q.Z. Development and Lactation Biology of Mammary Gland, 1st ed.; Science Press: Beijing, China, 2009; pp. 59–61. ISBN 978-7-03-023088-1. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Steinhardt, A.A.; Gayyed, M.F.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Lim, C.J.; Chen, L.; Chong, Y.F.; Huang, C.; Song, H.; Hong, W. The Hippo pathway in biological control and cancer development. J. Cell Physiol. 2011, 226, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef]
- Hao, Y.; Chun, A.; Cheung, K.; Rashidi, B.; Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008, 283, 5496–5509. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef]
- Goulev, Y.; Fauny, J.D.; Gonzalez-Marti, B.; Flagiello, D.; Silber, J.; Zider, A. SCALLOPED Interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008, 18, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, Y.; Zheng, Y.; Dong, J.; Pan, D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 2008, 14, 388–398. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, F.; Zhang, Q.; Chen, Y.; Wang, B.; Jiang, J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 2008, 14, 377–387. [Google Scholar] [CrossRef]
- Xu, C.; Wang, L.; Zhang, Y.; Li, W.; Li, J.; Wang, Y.; Meng, C.; Qin, J.; Zheng, Z.H.; Lan, H.Y.; et al. Tubule-Specific Mst1/2 Deficiency Induces CKD via YAP and Non-YAP Mechanisms. J. Am. Soc. Nephrol. 2020, 31, 946–961. [Google Scholar] [CrossRef]
- Tao, W.; Zhang, S.; Turenchalk, G.S.; Stewart, R.A.; St John, M.A.; Chen, W.; Xu, T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 1999, 21, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Hirota, T.; Morisaki, T.; Hara, T.; Marumoto, T.; Iida, S.; Makino, K.; Yamamoto, H.; Hiraoka, T.; Kitamura, N.; et al. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 1999, 459, 159–165. [Google Scholar] [CrossRef]
- Yabuta, N.; Fujii, T.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Nishiguchi, H.; Endo, Y.; Toji, S.; Tanaka, H.; Nishimune, Y.; et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics 2000, 63, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Takaori-Kondo, A.; Kamikubo, Y.; Uchiyama, T. Molecular cloning of a novel human protein kinase, kpm, that is homologous to warts/lats, a Drosophila tumor suppressor. Oncogene 2000, 19, 3101–3109. [Google Scholar] [CrossRef] [PubMed]
- St John, M.A.; Tao, W.; Fei, X.; Fukumoto, R.; Carcangiu, M.L.; Brownstein, D.G.; Parlow, A.F.; McGrath, J.; Xu, T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 1999, 21, 182–186. [Google Scholar] [CrossRef]
- Lee, K.M.; Yasuda, H.; Hollingsworth, M.A.; Ouellette, M.M. Notch 2-positive progenitors with the intrinsic ability to give rise to pancreatic ductal cells. Lab. Investig. 2005, 85, 1003–1012. [Google Scholar] [CrossRef]

| Group | Number of Yaks (n) | Crown–Rump Length (cm) | Characteristics |
|---|---|---|---|
| GP | 4 | 20, 26, 28, 30 | low milk exudation |
| LP | 4 | none | large milk exudation |
| DP | 4 | none | no milk exudation |
| Primer | Forward or Reverse | Sequence (5′–3′) | GenBank No. | Size |
|---|---|---|---|---|
| MST1 LATS1 YAP1 TEAD1 β-actin | Forward Reverse Forward Reverse Forward Reverse Forward Reverse Forward Reverse | GCTGCTTCTGATATGGTTCTC AGTCGTGTATGAGGTGAGTG TCCTCCACCACCTCTTAACAC TCTGACCGCTACCATTCTGAA GCTCTCCGTAGCCAGTTACCAA ATGCTGAGTCCGCTGTCTGTG GCCAACCATTCTTACAGTGA CTTGCGTATTCCGTCTCTAC CCGTGACATCAAGGAGAAG AGGAAGGAAGGCTGGAAG | NM_001075677.2 NM_001192866.1 XM_024975708.1 XM_024975797.1 DQ: 838049.1 | 245 bp 161 bp 173 bp 296 bp 174 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; Lu, B.; Zhao, L.; Yao, Y.; Qian, W.; Yu, Z.; Cui, Y.; Yu, S.; Fan, J. Expression of Key Factors of the Hippo Signaling Pathway in Yak (Bos grunniens) Mammary Gland. Animals 2022, 12, 2103. https://doi.org/10.3390/ani12162103
Du P, Lu B, Zhao L, Yao Y, Qian W, Yu Z, Cui Y, Yu S, Fan J. Expression of Key Factors of the Hippo Signaling Pathway in Yak (Bos grunniens) Mammary Gland. Animals. 2022; 12(16):2103. https://doi.org/10.3390/ani12162103
Chicago/Turabian StyleDu, Peiyan, Bin Lu, Ling Zhao, Ying Yao, Wenjie Qian, Zhen Yu, Yan Cui, Sijiu Yu, and Jiangfeng Fan. 2022. "Expression of Key Factors of the Hippo Signaling Pathway in Yak (Bos grunniens) Mammary Gland" Animals 12, no. 16: 2103. https://doi.org/10.3390/ani12162103
APA StyleDu, P., Lu, B., Zhao, L., Yao, Y., Qian, W., Yu, Z., Cui, Y., Yu, S., & Fan, J. (2022). Expression of Key Factors of the Hippo Signaling Pathway in Yak (Bos grunniens) Mammary Gland. Animals, 12(16), 2103. https://doi.org/10.3390/ani12162103





