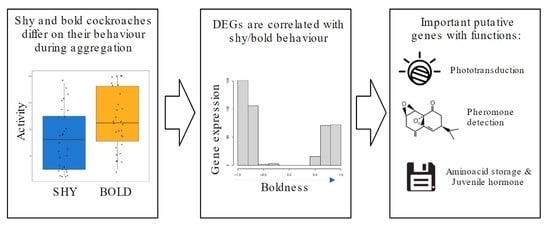
Differential Gene Expression Correlates with Behavioural Polymorphism during Collective Behaviour in Cockroaches
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Behavioural Experiments
2.2. Sample Preparation and RNA Sequencing
2.3. Differential Gene Expression Analysis
2.4. Gene Ontology Analysis
2.5. Statistical Analysis for Behavioural Data
3. Results
3.1. Behavioural Experiments
3.2. Differential Gene Expression
3.3. Gene Ontology Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Réale, D.; Dingemanse, N.J.; Kazem, A.J.N.; Wright, J. Evolutionary and ecological approaches to the study of personality. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3937–3946. [Google Scholar] [CrossRef]
- Carere, C.; Maestripieri, D. Animal Personalities: Behavior, Physiology, and Evolution; University of Chicago Press: Chicago, IL, USA, 2013. [Google Scholar]
- Wolf, M.; Weissing, F.J. Animal personalities: Consequences for ecology and evolution. Trends Ecol. Evol. 2012, 27, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Dall, S.R.X.; Houston, A.I.; McNamara, J.M. The behavioural ecology of personality: Consistent individual differences from an adaptive perspective. Ecol. Lett. 2004, 7, 734–739. [Google Scholar] [CrossRef]
- Sih, A.; Bell, A.; Johnson, J.C. Behavioral syndromes: An ecological and evolutionary overview. Trends Ecol. Evol. 2004, 19, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Hui, A.; Pinter-Wollman, N. Individual variation in exploratory behaviour improves speed and accuracy of collective nest selection by Argentine ants. Anim. Behav. 2014, 93, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Modlmeier, A.P.; Liebmann, J.E.; Foitzik, S. Diverse societies are more productive: A lesson from ants. Proc. R. Soc. B Boil. Sci. 2012, 279, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Nicolis, S.C.; Deneubourg, J.-L. The effect of idiosyncrasy on aggregation in group-living organisms. J. Theor. Biol. 2022, 542, 111120. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.R.; Seeley, T.D. Genetic Diversity in Honey Bee Colonies Enhances Productivity and Fitness. Science 2007, 317, 362–364. [Google Scholar] [CrossRef]
- Friedman, D.A.; York, R.A.; Hilliard, A.T.; Gordon, D.M. Gene expression variation in the brains of harvester ant foragers is associated with collective behavior. Commun. Biol. 2020, 3, 100. [Google Scholar] [CrossRef]
- Tang, W.; Davidson, J.D.; Zhang, G.; Conen, K.E.; Fang, J.; Serluca, F.; Li, J.; Xiong, X.; Coble, M.; Tsai, T.; et al. Genetic Control of Collective Behavior in Zebrafish. iScience 2020, 23, 100942. [Google Scholar] [CrossRef] [Green Version]
- Bell, A.M.; Aubin-Horth, N. What can whole genome expression data tell us about the ecology and evolution of personality? Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 4001–4012. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Pan, H.; Li, C.; Salzberg, S.L.; Puiu, D.; Magoc, T.; Robertson, H.M.; Hudson, M.E.; Venkat, A.; Fischman, B.J.; et al. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science 2015, 348, 1139–1143. [Google Scholar] [CrossRef]
- Kocher, S.D.; Mallarino, R.; Rubin, B.E.R.; Yu, D.W.; Hoekstra, H.E.; Pierce, N.E. The genetic basis of a social polymorphism in halictid bees. Nat. Commun. 2018, 9, 4338. [Google Scholar] [CrossRef]
- Kaur, R.; Stoldt, M.; Jongepier, E.; Feldmeyer, B.; Menzel, F.; Bornberg-Bauer, E.; Foitzik, S. Ant behaviour and brain gene expression of defending hosts depend on the ecological success of the intruding social parasite. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180192. [Google Scholar] [CrossRef]
- Ogawa, S.; Eng, V.; Taylor, J.; Lubahn, D.B.; Korach, K.S.; Pfaff, D.W. Roles of estrogen receptor-α gene expression in reproduction-related behaviors in female mice. Endocrinology 1998, 139, 5070–5081. [Google Scholar] [CrossRef]
- Zayed, A.; Robinson, G.E. Understanding the Relationship Between Brain Gene Expression and Social Behavior: Lessons from the Honey Bee. Annu. Rev. Genet. 2012, 46, 591–615. [Google Scholar] [CrossRef]
- Robinson, G.E.; Fernald, R.D.; Clayton, D.F. Genes and Social Behavior. Science 2008, 322, 896–900. [Google Scholar] [CrossRef]
- Linksvayer, T.A.; Johnson, B.R. Re-thinking the social ladder approach for elucidating the evolution and molecular basis of insect societies. Curr. Opin. Insect Sci. 2019, 34, 123–129. [Google Scholar] [CrossRef]
- Patalano, S.; Vlasova, A.; Wyatt, C.; Ewels, P.; Camara, F.; Ferreira, P.G.; Asher, C.L.; Jurkowski, T.P.; Segonds-Pichon, A.; Bachman, M.; et al. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. USA 2015, 112, 13970–13975. [Google Scholar] [CrossRef]
- Yaguchi, H.; Suzuki, R.; Matsunami, M.; Shigenobu, S.; Maekawa, K. Transcriptomic changes during caste development through social interactions in the termite Zootermopsis nevadensis. Ecol. Evol. 2019, 9, 3446–3456. [Google Scholar] [CrossRef] [Green Version]
- Kronauer, D.J.; Libbrecht, R. Back to the roots: The importance of using simple insect societies to understand the molecular basis of complex social life. Curr. Opin. Insect Sci. 2018, 28, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Rehan, S.M.; Toth, A.L. Climbing the social ladder: The molecular evolution of sociality. Trends Ecol. Evol. 2015, 30, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Shell, W.A.; Rehan, S.M. Social modularity: Conserved genes and regulatory elements underlie caste-antecedent behavioural states in an incipiently social bee. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191815. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Watanabe, Y.; Tin, M.M.Y.; Tsuji, K.; Mikheyev, A.S. Social dominance alters nutrition-related gene expression immediately: Transcriptomic evidence from a monomorphic queenless ant. Mol. Ecol. 2017, 26, 2922–2938. [Google Scholar] [CrossRef] [PubMed]
- Inward, D.; Beccaloni, G.; Eggleton, P. Death of an order: A comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol. Lett. 2007, 3, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Planas-Sitjà, I.; Deneubourg, J.-L.; Gibon, C.; Sempo, G. Group personality during collective decision-making: A multi-level approach. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142515. [Google Scholar] [CrossRef] [PubMed]
- Crall, J.D.; Souffrant, A.D.; Akandwanaho, D.; Hescock, S.D.; Callan, S.E.; Coronado, W.M.; Baldwin, M.W.; de Bivort, B.L. Social context modulates idiosyncrasy of behaviour in the gregarious cockroach Blaberus discoidalis. Anim. Behav. 2015, 111, 297–305. [Google Scholar] [CrossRef]
- Amé, J.-M.; Halloy, J.; Rivault, C.; Detrain, C.; Deneubourg, J.L. Collegial decision making based on social amplification leads to optimal group formation. Proc. Natl. Acad. Sci. USA 2006, 103, 5835–5840. [Google Scholar] [CrossRef]
- Laurent Salazar, M.-O.; Planas-Sitjà, I.; Deneubourg, J.-L.; Sempo, G. Collective resilience in a disturbed environment: Stability of the activity rhythm and group personality in Periplaneta americana. Behav. Ecol. Sociobiol. 2015, 69, 1879–1896. [Google Scholar] [CrossRef]
- Sempo, G.; Canonge, S.; Deneubourg, J.-L. From Aggregation to Dispersion: How Habitat Fragmentation Prevents the Emergence of Consensual Decision Making in a Group. PLoS ONE 2013, 8, e78951. [Google Scholar] [CrossRef] [Green Version]
- Canonge, S.; Deneubourg, J.-L.; Sempo, G. Group Living Enhances Individual Resources Discrimination: The Use of Public Information by Cockroaches to Assess Shelter Quality. PLoS ONE 2011, 6, e19748. [Google Scholar] [CrossRef]
- Lihoreau, M.; Costa, J.T.; Rivault, C. The social biology of domiciliary cockroaches: Colony structure, kin recognition and collective decisions. Insectes Sociaux 2012, 59, 445–452. [Google Scholar] [CrossRef]
- Saïd, I.; Costagliola, G.; Leoncini, I.; Rivault, C. Cuticular hydrocarbon profiles and aggregation in four Periplaneta species (Insecta: Dictyoptera). J. Insect Physiol. 2005, 51, 995–1003. [Google Scholar] [CrossRef]
- Stanley, C.R.; Mettke-Hofmann, C.; Preziosi, R.F. Personality in the cockroach Diploptera punctata: Evidence for stability across developmental stages despite age effects on boldness. PLoS ONE 2017, 12, e0176564. [Google Scholar] [CrossRef]
- Arican, C.; Bulk, J.; Deisig, N.; Nawrot, M.P. Cockroaches Show Individuality in Learning and Memory During Classical and Operant Conditioning. Front. Physiol. 2020, 10, 1539. [Google Scholar] [CrossRef]
- Planas-Sitjà, I.; Deneubourg, J.-L. The role of personality variation, plasticity and social facilitation in cockroach aggregation. Biol. Open 2018, 7, bio036582. [Google Scholar] [CrossRef]
- Laurent Salazar, M.-O.L.; Planas-Sitjà, I.; Sempo, G.; Deneubourg, J.-L. Individual Thigmotactic Preference Affects the Fleeing Behavior of the American Cockroach (Blattodea: Blattidae). J. Insect Sci. 2018, 18, 9. [Google Scholar] [CrossRef]
- Planas-Sitjà, I.; Nicolis, S.C.; Sempo, G.; Deneubourg, J.-L. The interplay between personalities and social interactions affects the cohesion of the group and the speed of aggregation. PLoS ONE 2018, 13, e0201053. [Google Scholar] [CrossRef]
- Planas-Sitjà, I. Personality variation improves collective decision-making in cockroaches. Behav. Process. 2020, 177, 104147. [Google Scholar] [CrossRef]
- Stamps, J.; Groothuis, T.G.G. The development of animal personality: Relevance, concepts and perspectives. Biol. Rev. Camb. Philos. Soc. 2010, 85, 301–325. [Google Scholar] [CrossRef] [Green Version]
- Lihoreau, M.; Brepson, L.; Rivault, C. The weight of the clan: Even in insects, social isolation can induce a behavioural syndrome. Behav. Process. 2009, 82, 81–84. [Google Scholar] [CrossRef]
- Pogson, M. Simulation of Invertebrate Aggregation Shows the Importance of Stable Personality over Diversity in Consensus Decision-Making. PLoS ONE 2016, 11, e0165082. [Google Scholar] [CrossRef]
- Chapman, B.B.; Thain, H.; Coughlin, J.; Hughes, W.O.H. Behavioural syndromes at multiple scales in Myrmica ants. Anim. Behav. 2011, 82, 391–397. [Google Scholar] [CrossRef]
- Réale, D.; Reader, S.M.; Sol, D.; McDougall, P.T.; Dingemanse, N.J. Integrating animal temperament within ecology and evolution. Biol. Rev. 2007, 82, 291–318. [Google Scholar] [CrossRef]
- Sih, A.; Del Giudice, M. Linking behavioural syndromes and cognition: A behavioural ecology perspective. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2762–2772. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Bullard, J.H.; Purdom, E.; Hansen, K.D.; Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Esser, D.; Geleijnse, J.M.; Matualatupauw, J.C.; Dower, J.I.; Kromhout, D.; Hollman, P.C.H.; Afman, L.A. Pure flavonoid epicatechin and whole genome gene expression profiles in circulating immune cells in adults with elevated blood pressure: A randomised double-blind, placebo-controlled, crossover trial. PLoS ONE 2018, 13, e0194229. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, T.; Mokry, M.; Micha, D.; Netelenbos, C.; Bravenboer, N.; Gilijamse, M.; Eekhoff, E.M.; de Vries, T.J. Activin-A Induces Early Differential Gene Expression Exclusively in Periodontal Ligament Fibroblasts from Fibrodysplasia Ossificans Progressiva Patients. Biomedicines 2021, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.M.; Cox, C.L.; McGlothlin, J.W.; Card, D.C.; Andrew, A.L.; Castoe, T.A. Hormonally Mediated Increases in Sex-Biased Gene Expression Accompany the Breakdown of Between-Sex Genetic Correlations in a Sexually Dimorphic Lizard. Am. Nat. 2017, 189, 315–332. [Google Scholar] [CrossRef]
- Chen, T.W.; Gan, R.-C.; Fang, Y.-K.; Chien, K.-Y.; Liao, W.-C.; Chen, C.-C.; Wu, T.H.; Chang, I.Y.-F.; Yang, C.; Huang, P.-J.; et al. FunctionAnnotator, a versatile and efficient web tool for non-model organism annotation. Sci. Rep. 2017, 7, 10430. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J. Gene Set Enrichment Analysis with topGO, R Package, 2.44.0; Bioconductor: Boston, MA, USA, 2021.
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- He, P.; Li, Z.-Q.; Zhang, Y.-F.; Chen, L.; Wang, J.; Xu, L.; Zhang, Y.N.; He, M. Identification of odorant-binding and chemosensory protein genes and the ligand affinity of two of the encoded proteins suggest a complex olfactory perception system in Periplaneta americana. Insect Mol. Biol. 2017, 26, 687–701. [Google Scholar] [CrossRef]
- Li, Z.-Q.; He, P.; Zhang, Y.-N.; Dong, S.-L. Molecular and Functional Characterization of Three Odorant-Binding Protein from Periplaneta americana. PLoS ONE 2017, 12, e0170072. [Google Scholar] [CrossRef]
- Wu, C.H.; Lee, M.F.; Liao, S.C.; Luo, S.F. Sequencing analysis of cDNA clones encoding the american cockroach Cr-PI allergens:Homology with insect hemolymph proteins. J. Biol. Chem. 1996, 271, 17937–17943. [Google Scholar] [CrossRef]
- Fowler, M.A.; Montell, C. Drosophila TRP channels and animal behavior. Life Sci. 2013, 92, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Yuan, Q.; Vogt, N.; Looger, L.L.; Jan, L.Y.; Jan, Y.N. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 2010, 468, 921–926. [Google Scholar] [CrossRef] [PubMed]
- French, A.S.; Meisner, S.; Liu, H.; Weckström, M.; Torkkeli, P.H. Transcriptome analysis and RNA interference of cockroach phototransduction indicate three opsins and suggest a major role for TRPL channels. Front. Physiol. 2015, 6, 207. [Google Scholar] [CrossRef]
- O’Shea-Wheller, T.A.; Hunt, E.R.; Sasaki, T. Functional Heterogeneity in Superorganisms: Emerging Trends and Concepts. Ann. Entomol. Soc. Am. 2021, 114, 562–574. [Google Scholar] [CrossRef]
- Durier, V.; Rivault, C. Exploitation of home range and spatial distribution of resources in German cockroaches (Dictyoptera: Blattellidae). J. Econ. Entomol. 2003, 96, 1832–1837. [Google Scholar] [CrossRef]
- Planas-Sitjà, I.; Laurent Salazar, M.O.; Sempo, G.; Deneubourg, J.L. Emigration dynamics of cockroaches under different disturbance regimes do not depend on individual personalities. Sci. Rep. 2017, 7, 44528. [Google Scholar] [CrossRef] [PubMed]
- Mindykowski, B.; Jaenicke, E.; Tenzer, S.; Cirak, S.; Schweikardt, T.; Schild, H.; Decker, H. Cockroach allergens Per a 3 are oligomers. Dev. Comp. Immunol. 2010, 34, 722–733. [Google Scholar] [CrossRef]
- Zhou, X.; Oi, F.M.; Scharf, M.E. Social exploitation of hexamerin: RNAi reveals a major caste-regulatory factor in termites. Proc. Natl. Acad. Sci. USA 2006, 103, 4499–4504. [Google Scholar] [CrossRef]
- Cameron, R.C.; Duncan, E.J.; Dearden, P.K. Biased gene expression in early honeybee larval development. BMC Genom. 2013, 14, 903. [Google Scholar] [CrossRef]
- Masuoka, Y.; Toga, K.; Nalepa, C.A.; Maekawa, K. A Crucial Caste Regulation Gene Detected by Comparing Termites and Sister Group Cockroaches. Genetics 2018, 209, 1225–1234. [Google Scholar] [CrossRef]
- Horsten Burmester, T. Evolution and function of the insect hexamerins. Eur. J. Entomol. 1999, 96, 213–225. [Google Scholar]
- Rivault, C. Role Des Facteurs Sociaux Sur L’expression De La Rythmicite Circadienne Dans Un Groupe Chez Pekiplaneta Americana (Dictyopt.). Behaviour 1981, 77, 23–43. [Google Scholar] [CrossRef]
- Rivault, C. Utilisation de l’Espace Dans un Groupe de Periplaneta americana. Behaviour 1981, 79, 239–253. [Google Scholar] [CrossRef]
- Biro, A.P.; Stamps, A.J. Are animal personality traits linked to life-history productivity? Trends Ecol. Evol. 2008, 23, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Withee, J.R.; Rehan, S.M. Social Aggression, Experience, and Brain Gene Expression in a Subsocial Bee. Integr. Comp. Biol. 2017, 57, 640–648. [Google Scholar] [CrossRef]
- Schal, C.; Gautier, J.-Y.; Bell, W.J. Behavioural Ecology of Cockroaches. Biol. Rev. 1984, 59, 209–254. [Google Scholar] [CrossRef]
- Dingemanse, N.J.; Barber, I.; Wright, J.; Brommer, J.E. Quantitative genetics of behavioural reaction norms: Genetic correlations between personality and behavioural plasticity vary across stickleback populations. J. Evol. Biol. 2012, 25, 485–496. [Google Scholar] [CrossRef]
- Moore, A.J. The inheritance of social dominance, mating behaviour and attractiveness to mates in male Nauphoeta cinerea. Anim. Behav. 1990, 39, 388–397. [Google Scholar] [CrossRef]
- Yahav, T.; Privman, E. A comparative analysis of methods for de novo assembly of hymenopteran genomes using either haploid or diploid samples. Sci. Rep. 2019, 9, 6480. [Google Scholar] [CrossRef]
- Legendre, F.; D’Haese, C.A.; Deleporte, P.; Pellens, R.; Whiting, M.F.; Schliep, K.; Grandcolas, P. The evolution of social behaviour in Blaberid cockroaches with diverse habitats and social systems: Phylogenetic analysis of behavioural sequences. Biol. J. Linn. Soc. 2014, 111, 58–77. [Google Scholar] [CrossRef] [Green Version]



| Name of the Protein | Upregulated in | Function | References |
|---|---|---|---|
| PameOBP1 | LRT | Odorant-binding protein; putative pheromone-binding protein; enriched expression in males | [59,60] |
| Cr-Pl/Per a 3 | LRT | Arylphorin storage protein, part of hexamerins; storage of amino acids; bind juvenile hormone; melanin generation | [61] |
| pTRP | SRT | Major role in phototransduction | [62,63,64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Planas-Sitjà, I.; Deneubourg, J.-L.; Lafontaine, D.L.J.; Wacheul, L.; Cronin, A.L. Differential Gene Expression Correlates with Behavioural Polymorphism during Collective Behaviour in Cockroaches. Animals 2022, 12, 2354. https://doi.org/10.3390/ani12182354
Planas-Sitjà I, Deneubourg J-L, Lafontaine DLJ, Wacheul L, Cronin AL. Differential Gene Expression Correlates with Behavioural Polymorphism during Collective Behaviour in Cockroaches. Animals. 2022; 12(18):2354. https://doi.org/10.3390/ani12182354
Chicago/Turabian StylePlanas-Sitjà, Isaac, Jean-Louis Deneubourg, Denis L. J. Lafontaine, Ludivine Wacheul, and Adam L. Cronin. 2022. "Differential Gene Expression Correlates with Behavioural Polymorphism during Collective Behaviour in Cockroaches" Animals 12, no. 18: 2354. https://doi.org/10.3390/ani12182354