Characterization of Gut Microbiome in the Mud Snail Cipangopaludina cathayensis in Response to High-Temperature Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Snail Preparation and Experimental Design
2.2. Sample Collection
2.3. DNA Extraction, Bacterial 16S rRNA Amplification, and Sequencing
2.4. Sequencing Data Processing
2.5. Data Analysis
3. Results
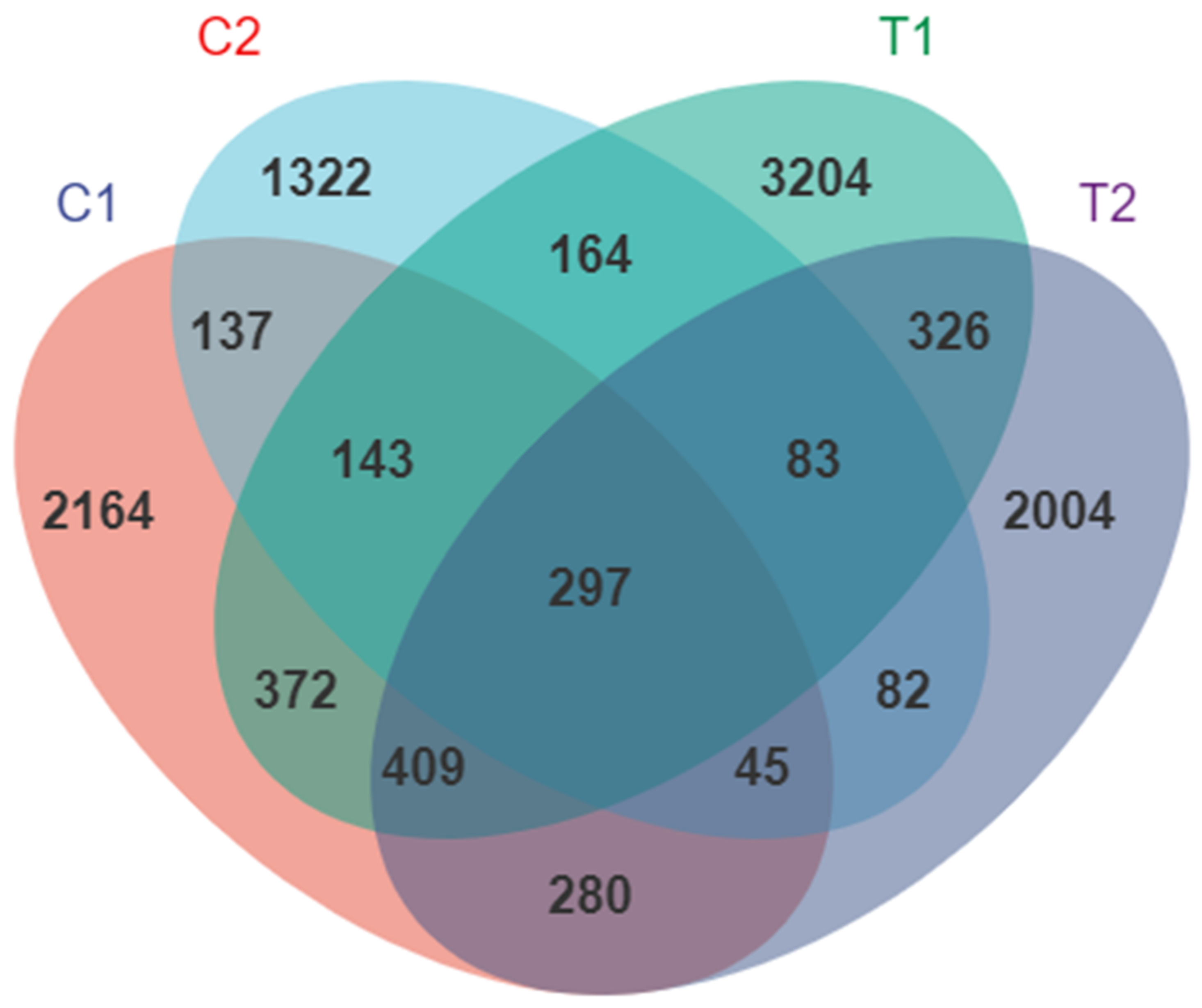
3.1. High-Throughput Sequencing Data
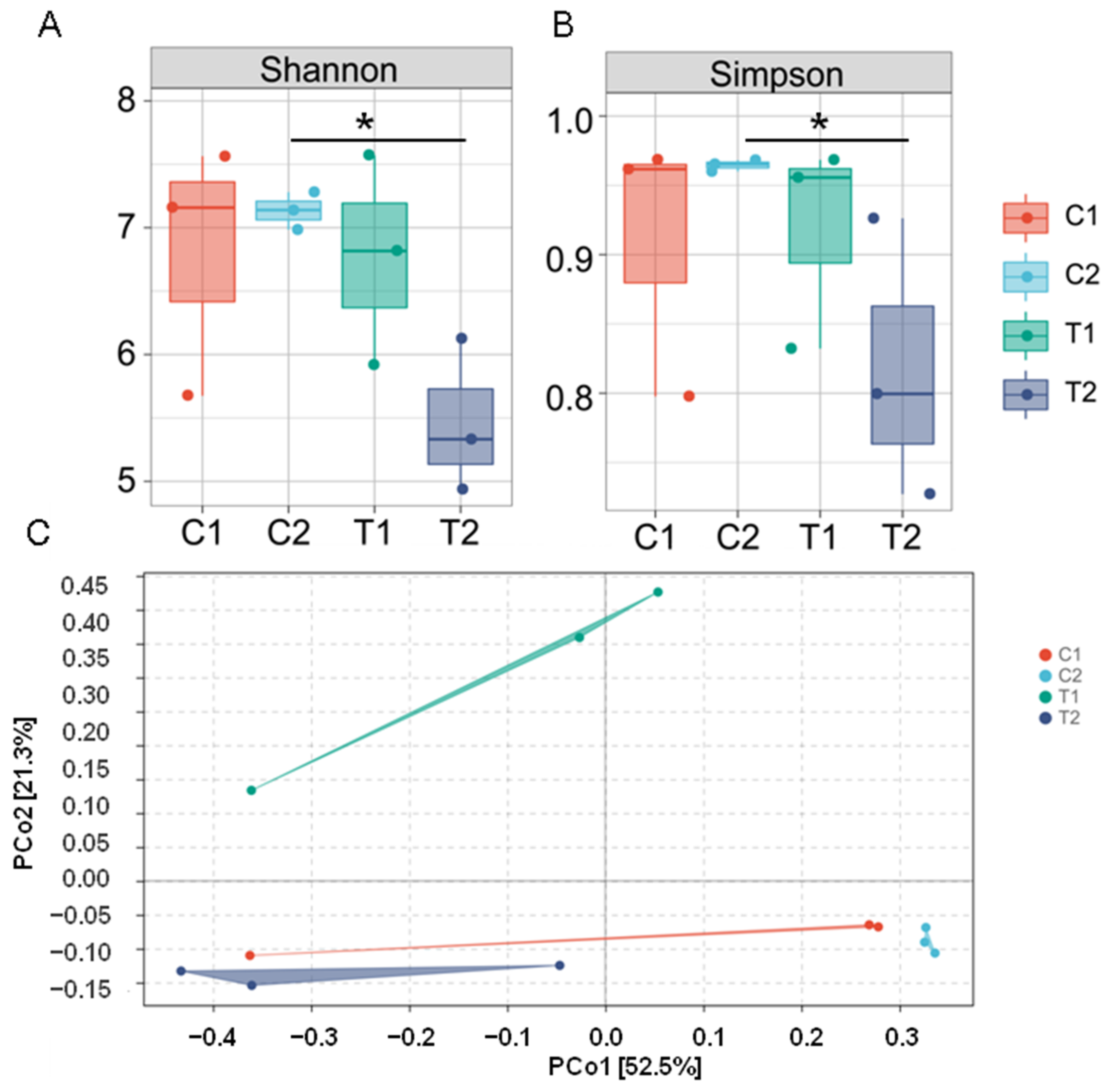
3.2. Diversity of Intestinal Microflora
3.3. Effect of High Temperature on the Overall Community Structure of Intestinal Microbiota
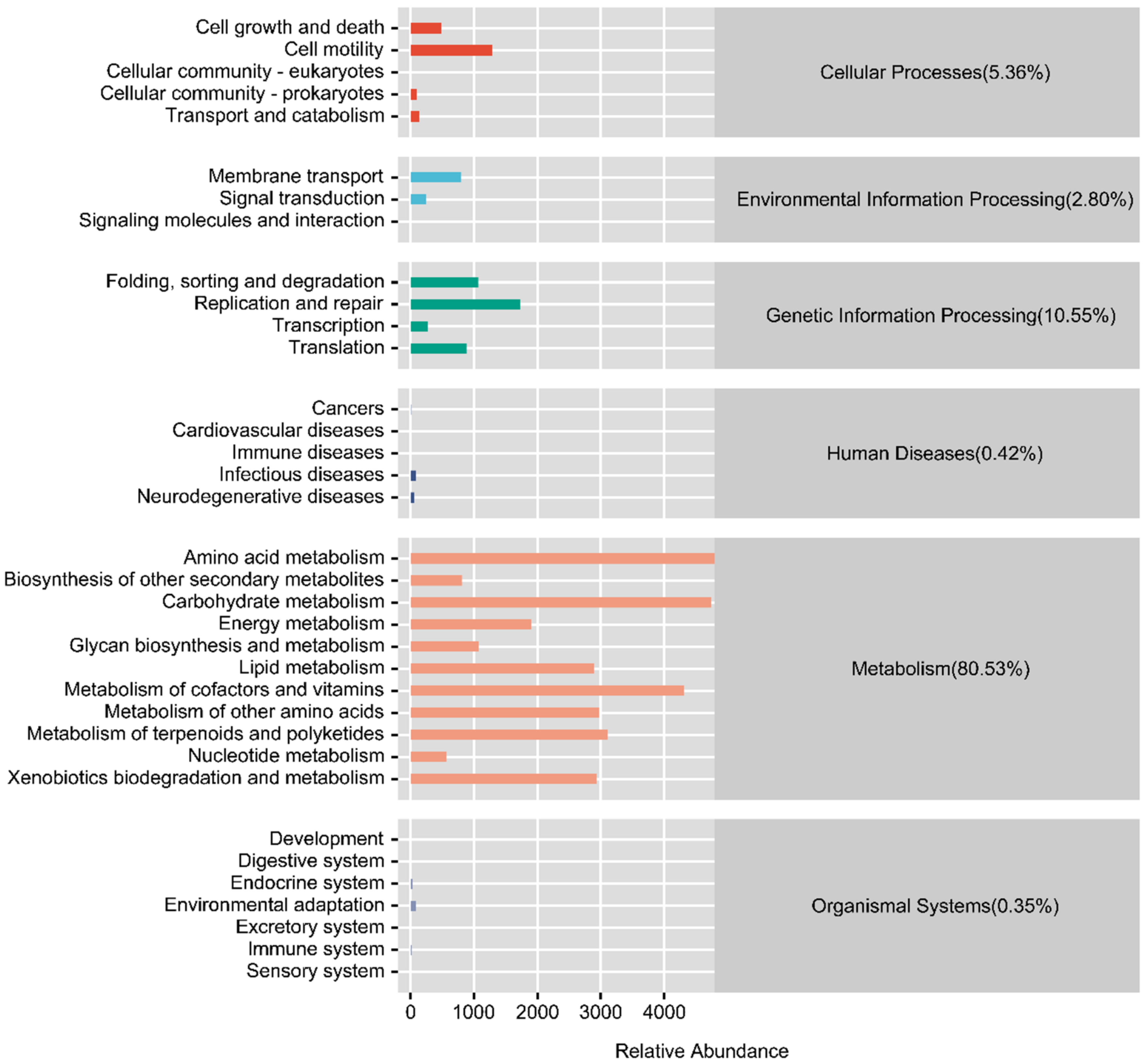
3.4. Functional Prediction of the Intestinal Microflora
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalon, P.W.; David, H.W. Growth and physiological responses in largemouth bass populations to environmental warming: Effects of inhabiting chronically heated environments. J. Therm. Biol. 2020, 88, 102467. [Google Scholar]
- Yang, T.Y.; Zhang, Y.; Meng, W.; Zhong, X.; Shan, Y.; Gao, T.X. Comparative transcriptomic analysis brings new insights into the response to acute temperature acclimation in burbot (Lota lota lota). Aquacult. Rep. 2021, 20, 100657. [Google Scholar] [CrossRef]
- Meredith, M.; Sommerkorn, M.; Cassotta, S.; Derksen, C.; Ekaykin, A.; Hollowed, A.; Kofinas, G.; Mackintosh, A.; Melbourne-Thomas, J.; Muelbert, M.M.C.; et al. (Eds.) IPCC Special Report on the Ocean and Cryosphere in a Changing Climate; IPCC: Geneva, Switzerland, 2019; Available online: https://www.ipcc.ch/srocc/chapter/chapter-3-2/ (accessed on 18 June 2022).
- Ahmed, N.; Thompson, S.; Glaser, M. Global aquaculture productivity, environmental sustainability, and climate change adaptability. Environ. Manag. 2019, 63, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.; Yu, X.B.; Wang, T.; Wang, Q.; Yao, W.Z.; Wu, Z.L. Combined effects of ration levels and temperature on immune responses of the triangle sail mussel Hyriopsis cumingii. Aquac. Res. 2022, 53, 440–452. [Google Scholar] [CrossRef]
- Zhao, T.T.; Ma, A.J.; Huang, Z.H.; Liu, Z.F.; Sun, Z.B.; Zhu, C.Y.; Yang, J.K.; Li, Y.D.; Wang, Q.M.; Qiao, X.W. Transcriptome analysis reveals that high temperatures alter modes of lipid metabolism in juvenile turbot (Scophthalmus maximus) liver. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jia, H.X.; Xiong, P.P.; Yao, G.Y.; He, M.X. Transcriptome and enzyme activity analyses of tolerance mechanisms in pearl oyster (Pinctada fucata) under high-temperature stress. Aquaculture 2022, 550, 737888. [Google Scholar] [CrossRef]
- Cannaday, J.D.; Farmer, T.M. Assessing the effects of sub-tropical winter thermal conditions on coolwater fish reproduction. Ecol. Freshw. Fish 2022, 3, 300–316. [Google Scholar] [CrossRef]
- Hue, T.; Chateau, O.; Lecellier, G.; Kayal, M.; Lanos, N.; Gossuin, H.; Adjeroud, M.; Dumas, P. Temperature affects the reproductive outputs of coral-eating starfish Acanthaster spp. after adult exposure to near-future ocean warming and acidification. Mar. Environ. Res. 2020, 162, 105164. [Google Scholar] [CrossRef]
- Watson, S.A.; Allan, B.J.M.; McQueen, D.E.; Nicol, S.; Parsons, D.M.; Pether, S.M.J.; Pope, S.; Setiawan, A.N.; Smith, N.; Wilson, C.; et al. Ocean warming has a greater effect than acidification on the early life history development and swimming performance of a large circumglobal pelagic fish. Glob. Chang. Biol. 2018, 24, 4368–4385. [Google Scholar] [CrossRef]
- Kong, H.; Clements, J.C.; Dupont, S.; Wang, T.; Huang, X.; Shang, Y.; Huang, W.; Chen, J.; Hu, M.; Wang, Y. Seawater acidification and temperature modulate anti-predator defenses in two co-existing Mytilus species. Mar. Pollut. Bull. 2019, 145, 118–125. [Google Scholar] [CrossRef]
- Lu, H.F.; Du, L.N.; Li, Z.Q.; Chen, X.Y.; Yang, J.X. Morphological analysis of the Chinese Cipangopaludina species (Gastropoda; Caenogastropoda: Viviparidae). Zool. Res. 2014, 35, 510–527. [Google Scholar]
- Luo, H.; Chen, L.T.; Jing, T.S.; Sun, W.B.; Li, Z.; Zhou, M.R.; Qin, J.Q.; Du, X.S.; Wen, L.T.; Pan, X.H.; et al. Muscle nutrition analysis of four snail species. J. Fish. China. 2021. in press. Available online: https://kns.cnki.net/kcms/detail/31.1283.S.20211018.1713.002.html (accessed on 20 June 2022).
- Zhao, T.; Xiong, J.Q.; Chen, W.; Xu, A.H.; Zhu, D.; Liu, J.T. Purification and characterization of a novel fibrinolytic enzyme from Cipangopaludina cahayensis. Iran. J. Biotechnol. 2021, 19, 121–127. [Google Scholar]
- Wang, C.; Liu, J.; Huang, Y.; Zhang, X. In vitro polysaccharide extraction from Cipangopaludina cathayensis and its pharmacological potential. J. Environ. Biol. 2016, 37, 1069–1072. [Google Scholar]
- Li, W.F.; Song, Y.J.; Wen, Y.H.; Luo, F.G.; Wang, W.M.; Li, Y.H. Investigation of current status of freshwater snail industry in China. Cult. Feed Sci. 2019, 11, 131–138. [Google Scholar]
- Woodward, G.; Perkins, D.M.; Brown, L.E. Climate change and freshwater ecosystems: Impacts across multiple levels of organization. Phil. Trans. R. Soc. B 2010, 365, 2093–2106. [Google Scholar] [CrossRef] [Green Version]
- Sangsawang, A.; Kovitvadhi, U.; Kovitvadhi, S. The effect of water temperature on the early-life development, growth and survival of the freshwater mussel Hyriopsis bialata. Aquaculture 2019, 510, 311–317. [Google Scholar] [CrossRef]
- Allan, E.R.O.; Blouin, M.S. Heat shock increases hydrogen peroxide release from circulating hemocytes of the snail Biomphalaria glabrata. Fish Shellfish Immun. 2020, 105, 203–208. [Google Scholar] [CrossRef]
- Hraoui, G.; Bettinazzi, S.; Gendron, A.D.; Boisclair, D.; Breton, S. Mitochondrial thermo-sensitivity in invasive and native freshwater mussels. J. Exp. Biol. 2020, 223, jeb215921. [Google Scholar] [CrossRef]
- Said, R.M.; Nassar, S.E. Mortality, energy reserves and oxidative stress responses of three native fresh water mussels to temperature as indicator of the potential impacts of climate change: A laboratory experiment approach. J. Therm. Biol. 2022, 104, 103154. [Google Scholar] [CrossRef]
- Sun, L.Y.; Wen, X.M.; Yu, N.; Chen, L.Q. Influence of water temperature and salinity on standard metabolism of Cipangopaludina cathayensis and Bellamya aeruginosa. J. Fish. Sci. China 2012, 19, 275–282. [Google Scholar] [CrossRef]
- Jiang, P.; Yu, X.J.; Yu, L.L.; Tian, F.W.; Zhao, J.X.; Zhang, H.; Chen, W.; Zhai, Q.X. The influence of gut microbiome on bone health and related dietary strategies against bone dysfunctions. Food Res. Int. 2021, 144, 110331. [Google Scholar]
- Talwar, C.; Nagar, S.; Lal, R.; Negi, R.K. Fish gut microbiome: Current approaches and future perspectives. Indian J. Microbiol. 2018, 58, 397–414. [Google Scholar] [CrossRef]
- Diwan, A.D.; Harke, S.N.; Gopalkrishna; Panche, A.N. Aquaculture industry prospective from gut microbiome of fish and shellfish: An overview. J. Anim. Physiol. Anim. Nutr. 2021, 106, 441–469. [Google Scholar] [CrossRef]
- Wang, N.; Jiang, M.; Zhang, P.J.; Shu, H.; Li, Y.R.; Guo, Z.Y.; Li, Y.H. Amelioration of Cd-induced bioaccumulation, oxidative stress and intestinal microbiota by Bacillus cereus in Carassius auratus gibelio. Chemosphere 2020, 245, 125613. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Zhou, J.; Li, J.Y.; Lv, W.; Zou, J.X.; Fan, L.F. A new insight into the intestine of pacific white shrimp: Regulation of intestinal homeostasis and regeneration in litopenaeus vannamei during temperature fluctuation. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100687. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Yang, N.; Liang, X.; Yoshida, A.; Osatomi, K.; Power, D.; Batista, F.M.; Yang, J.L. Elevated seawater temperatures decrease microbial diversity in the gut of Mytilus coruscus. Front. Physiol. 2018, 9, 839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.F.; Xu, J.K.; Chen, Y.W.; Ding, W.Y.; Shao, A.Q.; Liang, X.; Zhu, Y.T.; Yang, J.L. Characterization of gut microbiome in the mussel Mytilus galloprovincialis in response to thermal stress. Front. Physiol. 2019, 10, 1086. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Chen, Y.W.; Xu, J.K.; Ding, W.Y.; Shao, A.Q.; Zhu, Y.T.; Wang, C.; Liang, X.; Yang, J.L. Temperature elevation and Vibrio cyclitrophicus infection reduce the diversity of haemolymph microbiome of the mussel Mytilus coruscus. Sci. Rep. 2019, 9, 16391. [Google Scholar] [CrossRef]
- Huang, Z.J.; Zeng, S.Z.; Xiong, J.H.; Hou, D.W.; Zhou, R.J.; Xing, C.G.; Wei, D.D.; Deng, X.S.; Yu, L.F.; Wang, H.; et al. Microecological Koch’s postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 2020, 8, 32. [Google Scholar] [CrossRef]
- Kong, N.; Han, S.; Fu, Q.; Yu, Z.C.; Wang, L.L.; Song, L.S. Impact of ocean acidification on the intestinal microflora of the pacific oyster Crassostrea gigas. Aquaculture 2022, 546, 737365. [Google Scholar] [CrossRef]
- Chen, J.J.; Cao, J.L.; Li, X.; Yin, Q.L. Effects of fluoride on the activities of antioxidant enzymes and MDA levels in hepatopancreas of Cipangopaludina cahayensis. Asian J. Ecotoxicol. 2018, 13, 268–273. [Google Scholar]
- Jiang, J.J.; Li, W.H.; Wu, Y.Y.; Cheng, C.X.; Ye, Q.Q.; Feng, J.X.; Xie, Z.X. Effects of cadmium exposure on intestinal microflora of Cipangopaludina cathayensis. Front. Microbiol. 2022, 13, 984757. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v2. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecolo. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 2006, 9, 683–693. [Google Scholar] [CrossRef]
- Scanes, E.; Parker, L.M.; Seymour, J.R.; Siboni, N.; King, W.L.; Danckert, N.P.; Wegner, K.M.; Dove, M.C.; O’Connor, W.A.; Ross, P.M. Climate change alters the haemolymph microbiome of oysters. Mar. Pollut. Bull. 2021, 164, 111991. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.; Moeller, A.H. The Effects of temperature on animal gut microbiomes. Front. Microbiol. 2020, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; Tang, B.; Luo, X.; Ke, C.H.; Huang, M.Q.; You, W.W.; Wang, Y. Effects of temperature, diet and genotype-induced variations on the gut microbiota of abalone. Aquaculture 2020, 524, 735269. [Google Scholar] [CrossRef]
- Duan, Y.F.; Xiong, D.L.; Wang, Y.; Li, H.; Dong, H.B.; Zhang, J.S. Toxic effects of ammonia and thermal stress on the intestinal microbiota and transcriptomic and metabolomic responses of Litopenaeus vannamei. Sci. Total Environ. 2021, 754, 141867. [Google Scholar] [CrossRef]
- Musella, M.; Wathsala, R.; Tavella, T.; Rampelli, S.; Barone, M.; Palladino, G.; Biagi, E.; Brigidi, P.; Turroni, S.; Franzellitti, S.; et al. Tissue-scale microbiota of the Mediterranean mussel (Mytilus galloprovincialis) and its relationship with the environment. Sci. Total Environ. 2020, 717, 137209. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Wu, H.Y.; Li, D.H.; Zeng, W.L.; Huang, J.L.; Wu, Z.J. Comparison of gut microbiome in the Chinese mud snail (Cipangopaludina chinensis) and the invasive golden apple snail (Pomacea canaliculata). PeerJ 2022, 10, e13245. [Google Scholar] [CrossRef]
- Zhou, K.Q.; Qin, J.Q.; Pang, H.F.; Chen, Z.; Huang, Y.; Li, W.H.; Du, X.S.; Wen, L.T.; Pan, X.H.; Lin, Y. Comparison of the composition and function of gut microbes between adult and juvenile Cipangopaludina chinensis in the rice snail system. PeerJ 2022, 10, e13042. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Wang, Y.B.; Zhang, Z.; Ding, Q.W.; Yang, Y.L.; Olsen, R.E.; Ringo, E.; Bindelle, J.; Zhou, Z.G. Use of probiotics in aquaculture of china-a review of the past decade. Fish Shellfish Immunol. 2019, 86, 734–755. [Google Scholar] [CrossRef]
- Guo, Y.B.; Pan, Q.; Yan, S.Q.; Chen, Y.H.; Li, M.J.; Chen, D.; Han, H.C.; Wu, B.; Cai, J.P. Bdellovibrio and like organisms promoted growth and survival of juvenile abalone haliotis discus hannai Ino and modulated bacterial community structures in its gut. Aquacul. Int. 2017, 25, 1625–1643. [Google Scholar] [CrossRef]
- Hao, K.; Wu, Z.Q.; Li, D.L.; Yu, X.B.; Wang, G.X.; Ling, F. Effects of Dietary Administration of Shewanella xiamenensis A-1, Aeromonas veronii A-7, and Bacillus subtilis, Single or Combined, on the Grass Carp (Ctenopharyngodon idella) Intestinal Microbiota. Probiotics Antimicro. 2017, 9, 386–396. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Machiels, K.; Perrier, C.; Romero, C.; Maes, S.; Flahou, B.; Steppe, M.; Haesebrouck, F.; Sas, B.; Ducatelle, R.; et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 2013, 62, 1745–1752. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Cheng, J.X.; Xia, Y.Q.; Li, X.H.; Liu, Y.; Liu, P.F. Response mechanism of gut microbiome and metabolism of European seabass (Dicentrarchus labrax) to temperature stress. Sci. Total Environ. 2022, 813, 151786. [Google Scholar] [CrossRef]
- Grigoreva, I.N. Gallstone disease, obesity and the Firmicutes/Bacteroidetes ratio as a possible biomarker of gut dysbiosis. J. Pers. Med. 2021, 11, 13. [Google Scholar] [CrossRef]
- Chen, X.W.; Lu, D.Y.; Li, Z.H.; Yue, W.C.; Wang, J.; Jiang, X.Y.; Han, H.; Wang, C.H. Plant and animal-type feedstuff shape the gut microbiota and metabolic processes of the chinese mitten crab Eriocheir sinensis. Front. Vet. Sci. 2021, 8, 589624. [Google Scholar] [CrossRef]
- Jose, P.A.; Jha, B. Intertidal marine sediment harbours Actinobacteria with promising bioactive and biosynthetic potential. Sci. Rep. 2017, 7, 10041. [Google Scholar] [CrossRef]
- Rey, T.; Dumas, B. Plenty is no plague: Streptomyces symbiosis with crops. Trends Plant Sci. 2017, 22, 30–37. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef] [Green Version]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef]
- Sadeepa, D.; Sirisena, K.; Manage, P.M. Diversity of microbial communities in hot springs of Sri Lanka as revealed by 16S rRNA gene high-throughput sequencing analysis. Gene 2022, 812, 146103. [Google Scholar] [CrossRef]
- Tian, B.; Hua, Y. Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol. 2010, 18, 512–520. [Google Scholar] [CrossRef]
- Ren, C.L.; Teng, Y.R.; Chen, X.Y.; Shen, Y.J.; Xiao, H.; Wang, H.Y. Impacts of earthworm introduction and cadmium on microbial communities composition and function in soil. Environ. Toxicol. Phar. 2021, 83, 103606. [Google Scholar] [CrossRef] [PubMed]
- Cheaib, B.; Seghouani, H.; Ijaz, U.Z.; Derome, N. Community recovery dynamics in yellow perch microbiome after gradual and constant metallic perturbations. Microbiome 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zeng, X.H.; Jiang, N.; Zhou, Y.; Zeng, L.B. Pseudomonas alcaligenes infection and mortality in cultured Chinese sturgeon, Acipenser sinensis. Aquaculture 2015, 446, 37–41. [Google Scholar] [CrossRef]
- Fu, L.Q.; Zhang, X.P.; Wang, Y.B.; Peng, L.S.; Li, W.F. Nitrogen removal characteristics of Pseudomonas stutzeri F11 and its application in grass carp culture. Fish. Sci. 2017, 83, 89–98. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Jiang, C.; Ling, F.; Wang, G.X. Effects of dietary supplementation of intestinal autochthonous bacteria on the innate immunity and disease resistance of grass carp (Ctenopharyngodon idellus). Aquaculture 2015, 438, 105–114. [Google Scholar] [CrossRef]
- Ye, Q.; Feng, Y.Y.; Wang, Z.L.; Zhou, A.G.; Xie, S.L.; Fan, L.F.; Xiang, Q.; Song, E.F.; Zou, J.X. Effects of dietary Gelsemium elegans alkaloids on intestinal morphology, antioxidant status, immune responses and microbiota of Megalobrama amblycephala. Fish Shellfish Immunol. 2019, 94, 464–478. [Google Scholar] [CrossRef]
- Liao, Z.H.; Gong, Y.Y.; Zhao, W.; He, X.S.; Wei, D.; Niu, J. Comparison effect of Rhodobacter sphaeroides protein replace fishmeal on growth performance, intestinal morphology, hepatic antioxidant capacity and immune gene expression of Litopenaeus vannamei under low salt stress. Aquaculture 2022, 547, 737488. [Google Scholar] [CrossRef]
- Liu, H.S.; Li, X.; Lei, H.J.; Li, D.; Chen, H.X.; Schlenk, D.; Yan, B.; Luo, Y.J.; Xie, L.T. Dietary seleno-l-methionine alters the microbial communities and causes damage in the gastrointestinal tract of Japanese medaka Oryzias latipes. Environ. Sci. Technol. 2021, 55, 16515–16525. [Google Scholar] [CrossRef]
- Wang, X.H.; Hu, M.H.; Gu, H.H.; Zhang, L.B.; Shang, Y.Y.; Wang, T.; Wang, T.Y.; Zeng, J.N.; Ma, L.K.; Huang, W.; et al. Short-term exposure to norfloxacin induces oxidative stress, neurotoxicity and microbiota alteration in juvenile large yellow croaker Pseudosciaena crocea. Environ. Pollut. 2020, 267, 115397. [Google Scholar] [CrossRef]
- Liu, F.P.; Xu, X.F.; Chao, L.; Chen, K.; Shao, A.; Sun, D.Q.; Hong, Y.; Hu, R.J.; Jiang, P.; Zhang, N.; et al. Alteration of the gut microbiome in chronic kidney disease patients and its association with serum free immunoglobulin light chains. Front. Immunol. 2021, 12, 609700. [Google Scholar] [CrossRef]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Erturk-Hasdemir, D.; Kasper, D.L. Finding a needle in a haystack: Bacteroides fragilis polysaccharide A as the archetypical symbiosis factor. Ann. N. Y. Acad. Sci. 2018, 1417, 116–129. [Google Scholar] [CrossRef]
- Brown, E.M.; Ke, X.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 2019, 25, 668–680. [Google Scholar] [CrossRef]
- Zhou, Y.T.; Zhi, F.C. Lower level of Bacteroides in the gut microbiota is associated with inflammatory bowel disease: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 5828959. [Google Scholar] [CrossRef]
- Dou, G.M.; He, W.; Liu, H.C.; Ma, Y.C. Halomonas heilongjiangensis sp. nov., a novel moderately halophilic bacterium isolated from saline and alkaline soil. Antonie Van Leeuwenhoek 2015, 108, 403–413. [Google Scholar] [CrossRef]
- Yeo, S.H.; Kwak, J.H.; Kim, Y.U.; Lee, J.S.; Kim, H.J.; Park, K.H.; Lee, J.S.; Ha, G.Y.; Lee, J.H.; Lee, J.Y.; et al. Peritoneal dialysis-related peritonitis due to Halomonas hamiltonii A first case report. Medicine 2016, 95, e5424. [Google Scholar] [CrossRef]
- Yang, H.T.; Zou, S.S.; Zhai, L.J.; Wang, Y.; Zhang, F.M.; An, L.G.; Yang, G.W. Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 2017, 71, 35–42. [Google Scholar] [CrossRef]
- Ben Hamed, S.; Guardiola, F.; Morcillo, P.; Gonzalez-Parraga, P.; Tavares Ranzani-Paiva, M.J.; Esteban, M.A. Adhesion of pathogenic bacteria to polystyrene, skin and gut mucus of gilthead seabream, infectious capacity and antibiotics susceptibility. Bol. Inst. Pesca 2019, 45, e490. [Google Scholar]
- Jia, P.P.; Junaid, M.; Xin, G.Y.; Wang, Y.; Ma, Y.B.; Pei, D.S. Disruption of intestinal homeostasis through altered responses of the microbial community, energy metabolites, and immune system in Zebrafish after chronic exposure to DEHP. Front. Microbiol. 2021, 12, 729530. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.-Y.; Cheng, C.-X.; Yang, L.; Ye, Q.-Q.; Li, W.-H.; Jiang, J.-Y. Characterization of Gut Microbiome in the Mud Snail Cipangopaludina cathayensis in Response to High-Temperature Stress. Animals 2022, 12, 2361. https://doi.org/10.3390/ani12182361
Wu Y-Y, Cheng C-X, Yang L, Ye Q-Q, Li W-H, Jiang J-Y. Characterization of Gut Microbiome in the Mud Snail Cipangopaludina cathayensis in Response to High-Temperature Stress. Animals. 2022; 12(18):2361. https://doi.org/10.3390/ani12182361
Chicago/Turabian StyleWu, Yang-Yang, Chun-Xing Cheng, Liu Yang, Quan-Qing Ye, Wen-Hong Li, and Jiao-Yun Jiang. 2022. "Characterization of Gut Microbiome in the Mud Snail Cipangopaludina cathayensis in Response to High-Temperature Stress" Animals 12, no. 18: 2361. https://doi.org/10.3390/ani12182361
APA StyleWu, Y.-Y., Cheng, C.-X., Yang, L., Ye, Q.-Q., Li, W.-H., & Jiang, J.-Y. (2022). Characterization of Gut Microbiome in the Mud Snail Cipangopaludina cathayensis in Response to High-Temperature Stress. Animals, 12(18), 2361. https://doi.org/10.3390/ani12182361





