Applying Machine Learning Algorithms for the Classification of Mink Infected with Aleutian Disease Using Different Data Sources
Abstract
:Simple Summary
Abstract
1. Introduction
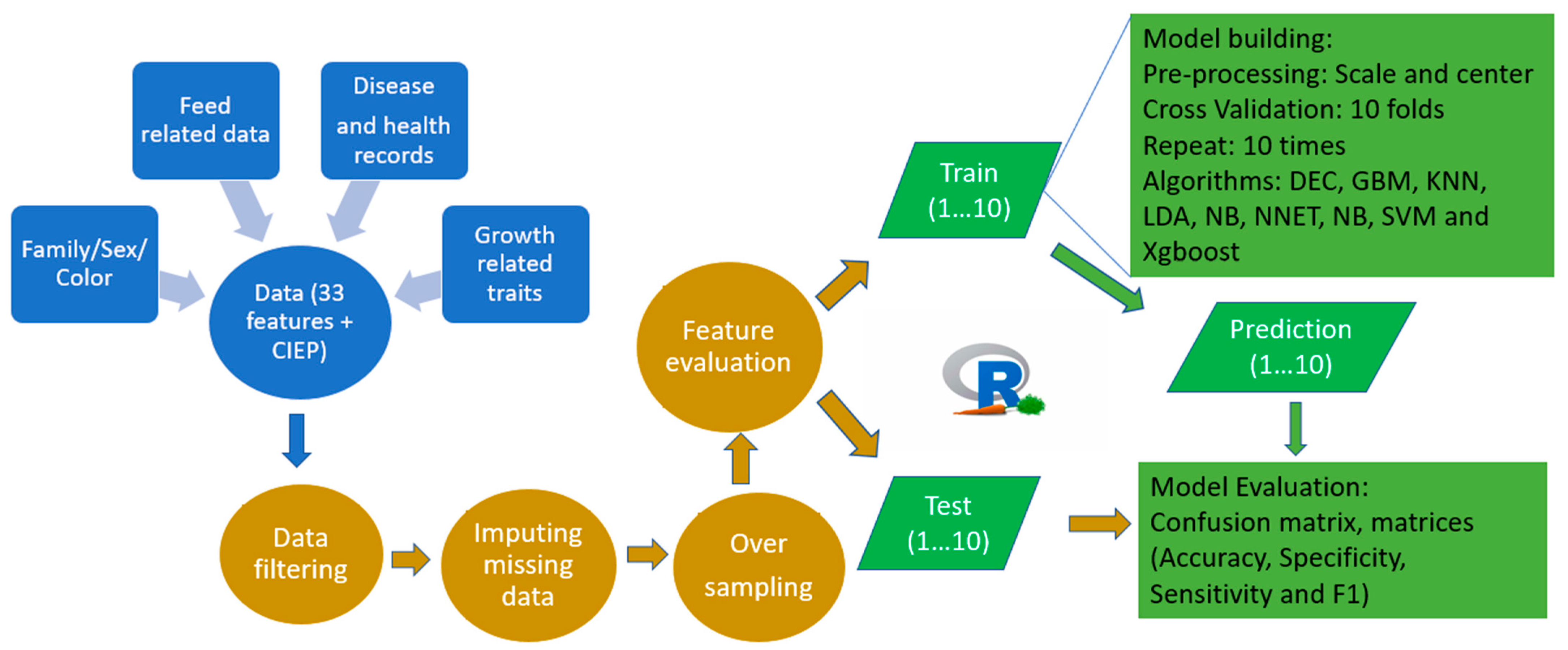
2. Materials and Methods
2.1. Animals and the Phenotypic Records
2.2. Algorithm Selection and the Data Preparation
2.3. Model Training and Performance Assessment
3. Results and Discussion
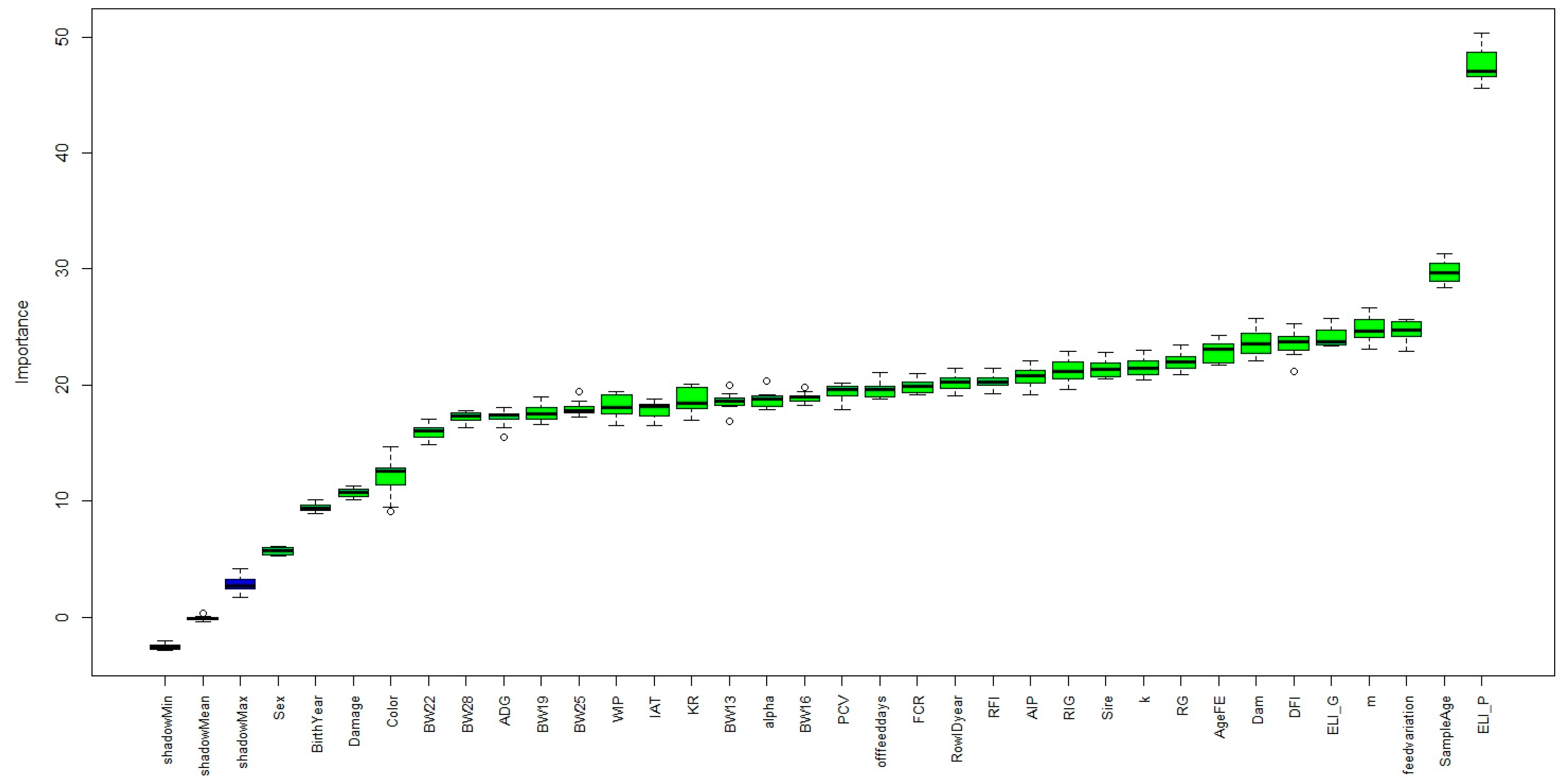
3.1. Feature Importance and the Model Performance
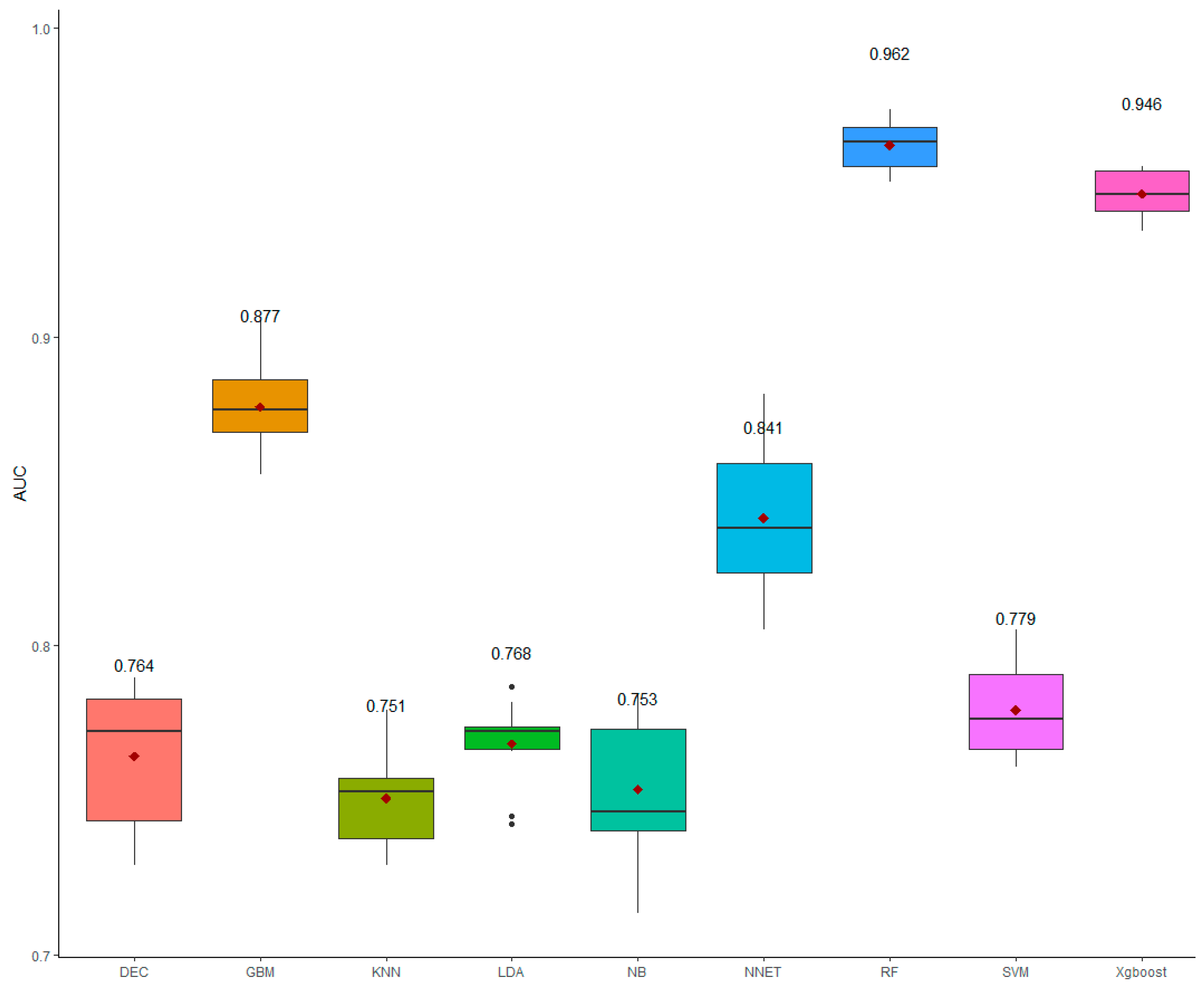
3.2. Performance Assessment
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manakhov, A.D.; Andreeva, T.V.; Trapezov, O.V.; Kolchanov, N.A.; Rogaev, E.I. Genome analysis identifies the mutant genes for common industrial Silverblue and Hedlund white coat colours in American mink. Sci. Rep. 2019, 9, 1–8. [Google Scholar]
- Hu, G.; Do, D.N.; Gray, J.; Miar, Y. Selection for favorable health traits: A potential approach to cope with diseases in farm animals. Animals 2020, 10, 1717. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.; Gorham, J.; Leader, R. Field test for Aleutian disease. Natl. Fur News 1962, 34, 8–9. [Google Scholar]
- Reichert, M.; Kostro, K. Effect of persistent infection of mink with Aleutian mink disease virus on reproductive failure. Bull. Vet. Inst. Pulawy. 2014, 58, 369–373. [Google Scholar] [CrossRef]
- Kowalczyk, M.; Gąsiorek, B.; Kostro, K.; Borzym, E.; Jakubczak, A. Breeding parameters on a mink farm infected with Aleutian mink disease virus following the use of methisoprinol. Arch. Virol. 2019, 164, 2691–2698. [Google Scholar] [CrossRef]
- Eklund, C.; Hadlow, W.; Kennedy, R.; Boyle, C.; Jackson, T. Aleutian disease of mink: Properties of the etiologic agent and the host responses. J. Infect. Dis. 1968, 118, 510–526. [Google Scholar] [CrossRef]
- Jensen, T.H.; Chriél, M.; Hansen, M.S. Progression of experimental chronic Aleutian mink disease virus infection. Acta Vet. Scand. 2016, 58, 35. [Google Scholar] [CrossRef]
- Farid, A.; Ferns, L. Aleutian mink disease virus infection may cause hair depigmentation. Scientifur 2011, 35, 55–59. [Google Scholar]
- Andersson, A.-M.; Wallgren, P. Evaluation of two enzyme-linked immunosorbent assays for serodiagnosis of Aleutian mink disease virus infection in mink. Acta Vet. Scand. 2013, 55, 1–6. [Google Scholar] [CrossRef]
- Christensen, L.S.; Gram-Hansen, L.; Chriél, M.; Jensen, T.H. Diversity and stability of Aleutian mink disease virus during bottleneck transitions resulting from eradication in domestic mink in Denmark. Vet. Microbiol. 2011, 149, 64–71. [Google Scholar] [CrossRef]
- Farid, A.H.; Zillig, M.L.; Finley, G.G.; Smith, G.C. Prevalence of the Aleutian mink disease virus infection in Nova Scotia, Canada. Prev. Vet. Med. 2012, 106, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, E. Documenting Freedom From Disease And Re-Establishing a Free Status After a Breakdown Aleutian Disease (Plasmacytosis) in Farmed Mink in Iceland. Acta Vet. Scand. 2001, 42, S87. [Google Scholar] [CrossRef]
- Themudo, G.E.; Østergaard, J.; Ersbøll, A.K. Persistent spatial clusters of plasmacytosis among Danish mink farms. Prev. Vet. Med. 2011, 102, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Knuuttila, A.; Aronen, P.; Eerola, M.; Gardner, I.A.; Virtala, A.-M.K.; Vapalahti, O. Validation of an automated ELISA system for detection of antibodies to Aleutian mink disease virus using blood samples collected in filter paper strips. Virol. J. 2014, 11, 141. [Google Scholar] [CrossRef]
- Sajda, P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 2006, 8, 537–565. [Google Scholar] [CrossRef]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The use of machine learning in rare diseases: A scoping review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine learning in agriculture: A review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Neethirajan, S. The role of sensors, big data and machine learning in modern animal farming. Sens. Bio-Sens. Res. 2020, 29, 100367. [Google Scholar] [CrossRef]
- Morota, G.; Ventura, R.V.; Silva, F.F.; Koyama, M.; Fernando, S.C. Big data analytics and precision animal agriculture symposium: Machine learning and data mining advance predictive big data analysis in precision animal agriculture. J. Anim. Sci. 2018, 96, 1540–1550. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Villalobos, N.; Vignes, M. Identifying Health Status in Grazing Dairy Cows from Milk Mid-Infrared Spectroscopy by Using Machine Learning Methods. Animals 2021, 11, 2154. [Google Scholar]
- Cairo, F.C.; Pereira, L.G.R.; Campos, M.M.; Tomich, T.R.; Coelho, S.G.; Lage, C.F.A.; Fonseca, A.P.; Borges, A.M.; Alves, B.R.C.; Dorea, J.R.R. Applying machine learning techniques on feeding behavior data for early estrus detection in dairy heifers. Comput. Electron. Agric. 2020, 179, 105855. [Google Scholar] [CrossRef]
- Alaiz-Rodríguez, R.; Parnell, A.C. A machine learning approach for lamb meat quality assessment using FTIR spectra. IEEE Access 2020, 8, 52385–52394. [Google Scholar] [CrossRef]
- Chen, D.; Wu, P.; Wang, K.; Wang, S.; Ji, X.; Shen, Q.; Yu, Y.; Qiu, X.; Xu, X.; Liu, Y. Combining computer vision score and conventional meat quality traits to estimate the intramuscular fat content using machine learning in pigs. Meat Sci. 2021, 185, 108727. [Google Scholar] [CrossRef]
- Taneja, M.; Byabazaire, J.; Jalodia, N.; Davy, A.; Olariu, C.; Malone, P. Machine learning based fog computing assisted data-driven approach for early lameness detection in dairy cattle. Comput. Electron. Agric. 2020, 171, 105286. [Google Scholar] [CrossRef]
- Kaler, J.; Mitsch, J.; Vázquez-Diosdado, J.A.; Bollard, N.; Dottorini, T.; Ellis, K.A. Automated detection of lameness in sheep using machine learning approaches: Novel insights into behavioural differences among lame and non-lame sheep. R. Soc. Open Sci. 2020, 7, 190824. [Google Scholar] [CrossRef] [PubMed]
- Fadul-Pacheco, L.; Delgado, H.; Cabrera, V.E. Exploring machine learning algorithms for early prediction of clinical mastitis. Int. Dairy J. 2021, 119, 105051. [Google Scholar] [CrossRef]
- Sun, Z.; Samarasinghe, S.; Jago, J. Detection of mastitis and its stage of progression by automatic milking systems using artificial neural networks. J. Dairy Res. 2010, 77, 168–175. [Google Scholar] [CrossRef]
- de Alencar Nääs, I.; da Silva Lima, N.D.; Gonçalves, R.F.; de Lima, L.A.; Ungaro, H.; Abe, J.M. Lameness prediction in broiler chicken using a machine learning technique. Inf. Process. Agric. 2021, 8, 409–418. [Google Scholar] [CrossRef]
- Mammadova, N.; Keskin, I. Application of the support vector machine to predict subclinical mastitis in dairy cattle. Sci. World J. 2013, 2013, 603897. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Miar, Y. Evaluation of growth curve models for body weight in American mink. Animals 2019, 10, 22. [Google Scholar]
- Hu, G.; Do, D.N.; Karimi, K.; Miar, Y. Genetic and phenotypic parameters for Aleutian disease tests and their correlations with pelt quality, reproductive performance, packed-cell volume, and harvest length in mink. J. Anim. Sci. 2021, 99, skab216. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Hu, G.; Salek Ardestani, S.; Miar, Y. Genetic and phenotypic parameters for body weights, harvest length, and growth curve parameters in American mink. J. Anim. Sci. 2021, 99, skab049. [Google Scholar] [CrossRef] [PubMed]
- Davoudi, P.; Do, D.N.; Colombo, S.; Rathgeber, B.; Hu, G.; Sargolzaei, M.; Wang, Z.; Plastow, G.; Miar, Y. Genetic and phenotypic parameters for feed efficiency and component traits in American mink. J. Anim. Sci. 2022, 100, skac216. [Google Scholar] [CrossRef] [PubMed]
- Bayrak, E.A.; Kırcı, P.; Ensari, T. Comparison of machine learning methods for breast cancer diagnosis. In Proceedings of the 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), Istanbul, Turkey, 24–26 April 2019; pp. 1–3. [Google Scholar]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.A.; Wishart, D.S. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2006, 2, 117693510600200030. [Google Scholar] [CrossRef]
- Bakoev, S.; Getmantseva, L.; Kolosova, M.; Kostyunina, O.; Chartier, D.R.; Tatarinova, T.V. PigLeg: Prediction of swine phenotype using machine learning. PeerJ 2020, 8, e8764. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Lunardon, N.; Menardi, G.; Torelli, N. ROSE: A package for binary imbalanced learning. R J. 2014, 6, 79–89. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Greiner, M.; Pfeiffer, D.; Smith, R. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 2000, 45, 23–41. [Google Scholar] [CrossRef]
- Knuuttila, A.; Aronen, P.; Saarinen, A.; Vapalahti, O. Development and Evaluation of an Enzyme-Linked Immunosorbent Assay Based on Recombinant VP2 Capsids for the Detection of Antibodies to Aleutian Mink Disease Virus. Clin. Vaccine Immunol. 2009, 16, 1360–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashtanov, S.; Salnikova, L. Aleutian mink disease: Epidemiological and genetic aspects. Biol. Bull. Rev. 2018, 8, 104–113. [Google Scholar] [CrossRef]
- Shao, Y.; Xiong, T.; Li, M.; Hayes, D.; Zhang, W.; Xie, W. China’s Missing Pigs: Correcting China’s Hog Inventory Data Using a Machine Learning Approach. Am. J. Agric. Econ. 2021, 103, 1082–1098. [Google Scholar] [CrossRef]
- Wang, X.; Shi, S.; Wang, G.; Luo, W.; Wei, X.; Qiu, A.; Luo, F.; Ding, X. Using Machine Learning To Improve the Accuracy of Genomic Prediction on Reproduction Traits in Pigs. J. Anim. Sci. Biotechnol. 2021, 13, 1–12. [Google Scholar]
- Chen, C.; Liaw, A.; Breiman, L. Using random forest to learn imbalanced data. Univ. Calif. Berkeley 2004, 110, 24. [Google Scholar]
- Balasso, P.; Marchesini, G.; Ughelini, N.; Serva, L.; Andrighetto, I. Machine learning to detect posture and behavior in dairy cows: Information from an accelerometer on the animal’s left flank. Animals 2021, 11, 2972. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
| CIEP Negative | CIEP Positive | ||||||
|---|---|---|---|---|---|---|---|
| Features | Name | N | Mean | SD | N | Mean | SD |
| General features | |||||||
| Sample_Age (day) | Age at the collection of blood for AD testing | 201 | 197.3 | 63.1 | 902 | 234.6 | 136.3 |
| RowIDyear | Row number where the mink were kept each year | 200 | 6.55 | 2.80 | 853 | 5.20 | 3.09 |
| AgeFE | Age of the mink (in days) when the feeding measure started | 200 | 198.25 | 3.50 | 852 | 197.60 | 9.73 |
| Damage | The damage score in the fur | 202 | 1.42 | 0.63 | 881 | 1.34 | 0.57 |
| Aleutian disease and health-related tests | |||||||
| Elisa_P | In vitro cultured Aleutian mink disease virus antigen-based enzyme-linked immunosorbent assay test | 203 | 0.33 | 0.97 | 924 | 1.67 | 2.29 |
| Elisa_G | Capsid protein of Aleutian mink disease virus-based enzyme-linked immunosorbent assay test | 203 | 0.64 | 0.98 | 922 | 2.46 | 2.19 |
| PCV | Packed cell volume | 201 | 58.11 | 2.94 | 919 | 56.67 | 4.03 |
| IAT | Iodine agglutination test | 200 | 0.42 | 0.60 | 918 | 0.77 | 1.05 |
| Feed intake and efficiency | |||||||
| DFI | Daily feed intake | 200 | 221.36 | 56.24 | 853 | 227.21 | 57.45 |
| ADG | Average Daily Gain | 199 | 7.45 | 3.40 | 821 | 8.60 | 3.86 |
| FCR | Food Conversion Ratio | 199 | 33.59 | 11.17 | 813 | 30.42 | 11.40 |
| RFI | Residual feed intake | 198 | 7.45 | 34.69 | 820 | −1.14 | 36.96 |
| RG | Residual intake and gain | 199 | 0.11 | 1.12 | 817 | −0.01 | 1.44 |
| RIG | Residual gain | 198 | −0.14 | 1.37 | 819 | 0.01 | 1.56 |
| KR | Kleiber ratio | 199 | 5.19 | 1.40 | 821 | 5.51 | 1.58 |
| Offfeeddays | Proportion of off-feed days based on feed intake | 199 | 0.05 | 0.09 | 842 | 0.06 | 0.08 |
| Feedvariation | Day-to-day variation in feed intake | 199 | 48.22 | 11.55 | 842 | 49.15 | 18.02 |
| Body weight and growth parameters | |||||||
| BW13 | Body weight at week 13 | 198 | 1.15 | 0.26 | 837 | 1.27 | 0.30 |
| BW16 | Body weight at week 16 | 198 | 1.42 | 0.36 | 834 | 1.58 | 0.41 |
| BW19 | Body weight at week 19 | 198 | 1.62 | 0.46 | 829 | 1.82 | 0.53 |
| BW22 | Body weight at week 22 | 198 | 1.82 | 0.54 | 824 | 2.04 | 0.61 |
| BW25 | Body weight at week 25 | 198 | 1.88 | 0.56 | 810 | 2.13 | 0.64 |
| BW28 | Body weight at week 28 | 197 | 1.92 | 0.59 | 805 | 2.17 | 0.67 |
| Alpha | Weight at maturity [34] | 198 | 1.99 | 0.63 | 807 | 2.25 | 0.71 |
| k | Maturation rate [34] | 198 | 0.24 | 0.10 | 803 | 0.24 | 0.11 |
| m | Inflection parameter [34] | 198 | 0.68 | 0.90 | 798 | 0.64 | 0.83 |
| AIP | Age at the inflection point [34] | 195 | 10.79 | 1.86 | 799 | 10.97 | 1.83 |
| WIP | Weight at the inflection point [34] | 196 | 0.89 | 0.33 | 804 | 1.01 | 0.35 |
| Algorithms | Sensitivity | Specificity | F1 | Accuracy |
|---|---|---|---|---|
| Artificial Neural Networks | 0.805 ± 0.008 | 0.877 ± 0.016 | 0.836 ± 0.096 | 0.841 ± 0.007 |
| Decision tree | 0.634 ± 0.007 | 0.894 ± 0.014 | 0.726 ± 0.01 | 0.764 ± 0.007 |
| Extreme Gradient Boosting | 0.905 ± 0.002 | 0.987 ± 0.005 | 0.944 ± 0.002 | 0.946 ± 0.002 |
| Gradient Boosting | 0.831 ± 0.005 | 0.924 ± 0.01 | 0.871 ± 0.035 | 0.877 ± 0.004 |
| K-Nearest Neighbors | 0.588 ± 0.006 | 0.909 ± 0.011 | 0.700 ± 0.000 | 0.749 ± 0.005 |
| Linear Discriminant Analysis | 0.672 ± 0.005 | 0.865 ± 0.009 | 0.743 ± 0.001 | 0.768 ± 0.004 |
| Naive Bayes | 0.666 ± 0.007 | 0.841 ± 0.014 | 0.73 ± 0.002 | 0.753 ± 0.007 |
| Random Forest | 0.938 ± 0.003 | 0.986 ± 0.005 | 0.961 ± 0.088 | 0.962 ± 0.002 |
| Support Vector Machines | 0.687 ± 0.005 | 0.872 ± 0.01 | 0.757 ± 0.001 | 0.779 ± 0.004 |
| Actual Data | ||||
| Positive | Negative | |||
| Predicted data | Positive | 184 | 4 | |
| Negative | 6 | 186 | ||
| Total | 190 | 190 | ||
| ModelA * | ModelB | FDR ** | ModelA | ModelB | FDR |
|---|---|---|---|---|---|
| GBM | DEC | 7.90 × 10−18 | RF | KNN | 7.90 × 10−18 |
| KNN | DEC | 2.36 × 10−3 | SVM | KNN | 1.18 × 10−7 |
| LDA | DEC | 5.15 × 10−3 | XGBOOST | KNN | 7.90 × 10−18 |
| NB | DEC | 8.90 × 10−6 | NB | LDA | 9.33 × 10−3 |
| NNET | DEC | 1.91 × 10−17 | NNET | LDA | 1.10 × 10−17 |
| RF | DEC | 7.90 × 10−18 | RF | LDA | 7.90 × 10−18 |
| SVM | DEC | 1.18 × 10−4 | SVM | LDA | 1.51 × 10−12 |
| XGBOOST | DEC | 7.90 × 10−18 | XGBOOST | LDA | 7.90 × 10−18 |
| KNN | GBM | 7.90 × 10−18 | NNET | NB | 1.10 × 10−17 |
| LDA | GBM | 7.90 × 10−18 | RF | NB | 7.90 × 10−18 |
| NB | GBM | 7.90 × 10−18 | SVM | NB | 8.91 × 10−12 |
| NNET | GBM | 9.37 × 10−13 | XGBOOST | NB | 7.90 × 10−18 |
| RF | GBM | 7.90 × 10−18 | RF | NNET | 7.90 × 10−18 |
| SVM | GBM | 7.90 × 10−18 | SVM | NNET | 2.23 × 10−17 |
| XGBOOST | GBM | 7.90 × 10−18 | XGBOOST | NNET | 7.90 × 10−18 |
| LDA | KNN | 0.18 | SVM | RF | 7.90 × 10−18 |
| NB | KNN | 0.51 | XGBOOST | RF | 3.77 × 10−8 |
| NNET | KNN | 1.08 × 10−17 | XGBOOST | SVM | 7.90 × 10−18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, D.N.; Hu, G.; Davoudi, P.; Shirzadifar, A.; Manafiazar, G.; Miar, Y. Applying Machine Learning Algorithms for the Classification of Mink Infected with Aleutian Disease Using Different Data Sources. Animals 2022, 12, 2386. https://doi.org/10.3390/ani12182386
Do DN, Hu G, Davoudi P, Shirzadifar A, Manafiazar G, Miar Y. Applying Machine Learning Algorithms for the Classification of Mink Infected with Aleutian Disease Using Different Data Sources. Animals. 2022; 12(18):2386. https://doi.org/10.3390/ani12182386
Chicago/Turabian StyleDo, Duy Ngoc, Guoyu Hu, Pourya Davoudi, Alimohammad Shirzadifar, Ghader Manafiazar, and Younes Miar. 2022. "Applying Machine Learning Algorithms for the Classification of Mink Infected with Aleutian Disease Using Different Data Sources" Animals 12, no. 18: 2386. https://doi.org/10.3390/ani12182386
APA StyleDo, D. N., Hu, G., Davoudi, P., Shirzadifar, A., Manafiazar, G., & Miar, Y. (2022). Applying Machine Learning Algorithms for the Classification of Mink Infected with Aleutian Disease Using Different Data Sources. Animals, 12(18), 2386. https://doi.org/10.3390/ani12182386









