“Guess Who’s Coming to Dinner”: Molecular Tools to Reconstruct multilocus Genetic Profiles from Wild Canid Consumption Remains
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
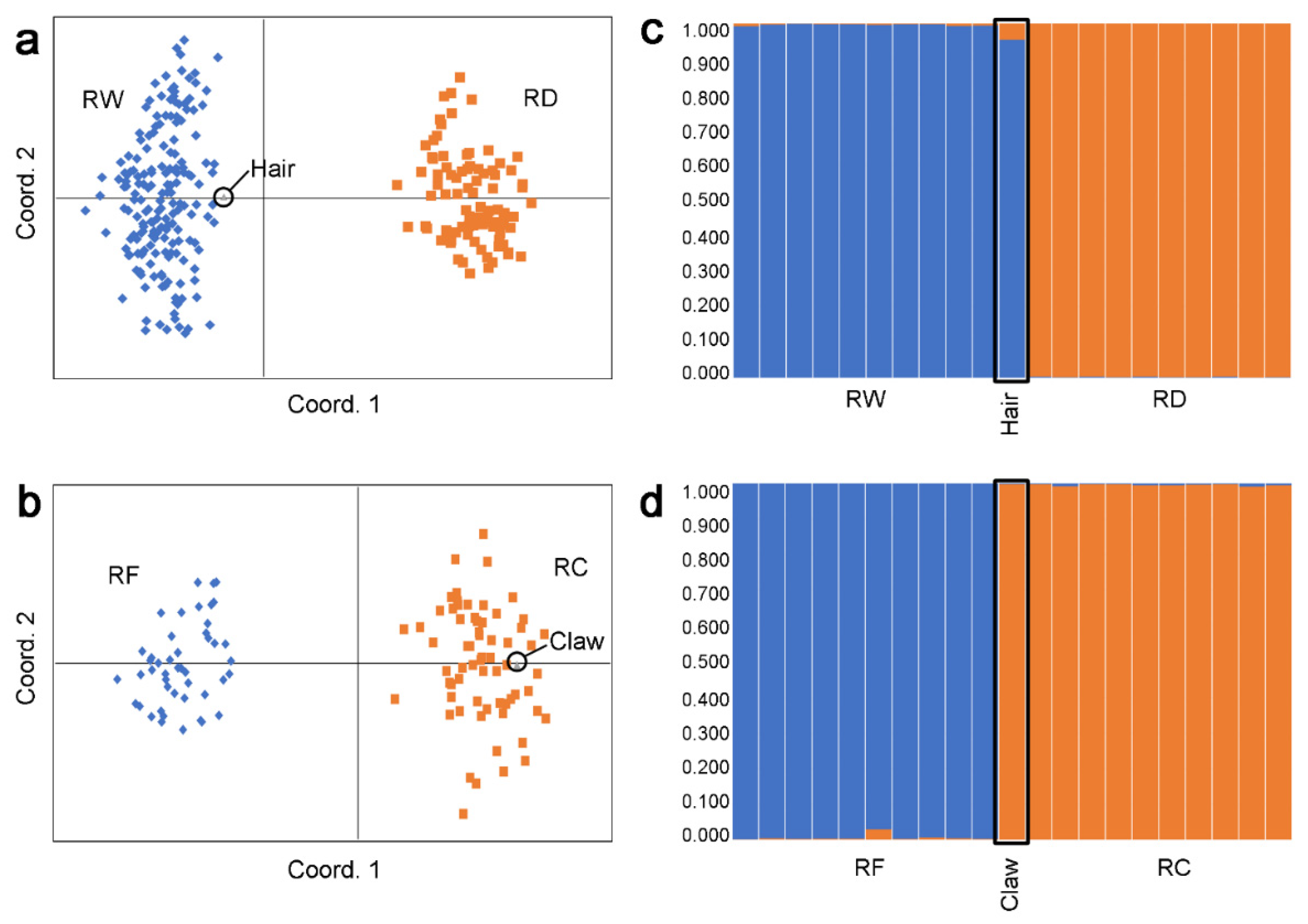
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trites, A.W.; Joy, R. Dietary Analysis from Fecal Samples: How Many Scats Are Enough? J. Mammal. 2005, 86, 704–712. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Chen, P.A.; Coverdale, T.C.; Erickson, D.L.; Kress, W.J.; Kuzmina, M.L.; Rubenstein, D.I.; Wang, W.; Pringle, R.M. DNA Metabarcoding Illuminates Dietary Niche Partitioning by African Large Herbivores. Proc. Natl. Acad. Sci. USA 2015, 112, 8019–8024. [Google Scholar] [CrossRef] [PubMed]
- Power, M.E.; Tilman, D.; Estes, J.A.; Menge, B.A.; Bond, W.J.; Mills, L.S.; Daily, G.; Castilla, J.C.; Lubchenco, J.; Paine, R.T. Challenges in the Quest for Keystones: Identifying Keystone Species Is Difficult—but Essential to Understanding How Loss of Species Will Affect Ecosystems. Bioscience 1996, 46, 609–620. [Google Scholar] [CrossRef]
- Chetri, M.; Odden, M.; Wegge, P. Snow Leopard and Himalayan Wolf: Food Habits and Prey Selection in the Central Himalayas, Nepal. PLoS ONE 2017, 12, e0170549. [Google Scholar] [CrossRef] [PubMed]
- Momeni, S.; Malekian, M.; Hemami, M.R. Molecular versus Morphological Approaches to Diet Analysis of the Caracal (Caracal caracal). Mammalia 2019, 83, 586–592. [Google Scholar] [CrossRef]
- Shehzad, W.; Riaz, T.; Nawaz, M.A.; Miquel, C.; Poillot, C.; Shah, S.A.; Pompanon, F.; Coissac, E.; Taberlet, P. Carnivore Diet Analysis Based on Next-Generation Sequencing: Application to the Leopard Cat (Prionailurus bengalensis) in Pakistan. Mol. Ecol. 2012, 21, 1951–1965. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Hoareau, Y.; Reese, E.; Wasser, S.K. Prey Partitioning between Sympatric Canid Species Revealed by DNA Metabarcoding. bioRxiv 2019, 22, 293–305. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.C.; Brown, D.S.; Jarman, S.N.; Taberlet, P. Who Is Eating What: Diet Assessment Using next Generation Sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef]
- Lee, O.; Lee, S.; Nam, D.H.; Lee, H.Y. Molecular Analysis for Investigating Dietary Habits: Genetic Screening of Prey Items In Scat And Stomach Contents Of Leopard Cats Prionailurus bengalensis euptilurus. Zool. Stud. 2013, 52, 45. [Google Scholar] [CrossRef]
- Bassi, E.; Canu, A.; Firmo, I.; Mattioli, L.; Scandura, M.; Apollonio, M. Trophic Overlap between Wolves and Free-Ranging Wolf x Dog Hybrids in the Apennine Mountains, Italy. Glob. Ecol. Conserv. 2017, 9, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Randi, E.; Pierpaoli, M.; Beaumont, M.; Ragni, B.; Sforzi, A. Genetic Identification of Wild and Domestic Cats (Felis silvestris) and Their Hybrids Using Bayesian Clustering Methods. Mol. Biol. Evol. 2001, 18, 1679–1693. [Google Scholar] [CrossRef] [PubMed]
- Randi, E.; Hulva, P.; Fabbri, E.; Galaverni, M.; Galov, A.; Kusak, J.; Bigi, D.; Bolfíková, B.Č.; Smetanová, M.; Caniglia, R.; et al. Multilocus Detection of Wolf x Dog Hybridization in Italy, and Guidelines for Marker Selection. PLoS ONE 2014, 9, e86409. [Google Scholar] [CrossRef] [PubMed]
- Galaverni, M.; Caniglia, R.; Pagani, L.; Fabbri, E.; Boattini, A.; Randi, E. Disentangling Timing of Admixture, Patterns of Introgression, and Phenotypic Indicators in a Hybridizing Wolf Population. Mol. Biol. Evol. 2017, 34, 2324–2339. [Google Scholar] [CrossRef]
- Mattucci, F.; Oliveira, R.; Lyons, L.A.; Alves, P.C.; Randi, E. European Wildcat Populations Are Subdivided into Five Main Biogeographic Groups: Consequences of Pleistocene Climate Changes or Recent Anthropogenic Fragmentation? Ecol. Evol. 2016, 6, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Galaverni, M.; Caniglia, R.; Fabbri, E.; Milanesi, P.; Randi, E. One, No One, or One Hundred Thousand: How Many Wolves Are There Currently in Italy? Mammal Res. 2016, 61, 13–24. [Google Scholar] [CrossRef]
- Velli, E.; Bologna, M.A.; Silvia, C.; Ragni, B.; Randi, E. Non-Invasive Monitoring of the European Wildcat (Felis silvestris silvestris Schreber, 1777): Comparative Analysis of Three Different Monitoring Techniques and Evaluation of Their Integration. Eur. J. Wildl. Res. 2015, 61, 657–668. [Google Scholar] [CrossRef]
- Caniglia, R.; Fabbri, E.; Galaverni, M.; Milanesi, P.; Randi, E. Non-invasive Sampling and Genetic Variability, Pack Structure, and Dynamics in an Expanding Wolf Population. J. Mammal. 2014, 95, 41–59. [Google Scholar] [CrossRef]
- Imbert, C.; Caniglia, R.; Fabbri, E.; Milanesi, P.; Randi, E.; Serafini, M.; Torretta, E.; Meriggi, A. Why Do Wolves Eat Livestock?: Factors Influencing Wolf Diet in Northern Italy. Biol. Conserv. 2016, 195, 156–168. [Google Scholar] [CrossRef]
- Lozano, J.; Malo, A.F. Conservation of The European Wildcat (Felis silvestris) In Mediterranean Environments: A Reassessment Of Current Threats. In Mediterranean Ecosystems: Dynamics, Management and Conservation; Williams, G.S., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2012; pp. 1–31. [Google Scholar]
- Mattucci, F.; Galaverni, M.; Lyons, L.A.; Alves, P.C.; Randi, E.; Velli, E.; Pagani, L.; Caniglia, R. Genomic Approaches to Identify Hybrids and Estimate Admixture Times in European Wildcat Populations. Sci. Rep. 2019, 9, 11612. [Google Scholar] [CrossRef]
- Caniglia, R.; Galaverni, M.; Velli, E.; Mattucci, F.; Canu, A.; Apollonio, M.; Mucci, N.; Scandura, M.; Fabbri, E. A Standardized Approach to Empirically Define Reliable Assignment Thresholds and Appropriate Management Categories in Deeply Introgressed Populations. Sci. Rep. 2020, 10, 2862. [Google Scholar] [CrossRef] [Green Version]
- Ferretti, F.; Lovari, S.; Mancino, V.; Burrini, L.; Rossa, M. Food Habits of Wolves and Selection of Wild Ungulates in a Prey-Rich Mediterranean Coastal Area. Mamm. Biol. 2019, 99, 119–127. [Google Scholar] [CrossRef]
- Figueiredo, A.M.; Valente, A.M.; Barros, T.; Carvalho, J.; Silva, D.A.M.; Fonseca, C.; de Carvalho, L.M.; Torres, R.T. What Does the Wolf Eat? Assessing the Diet of the Endangered Iberian Wolf (Canis Lupus signatus) in Northeast Portugal. PLoS ONE 2020, 15, e0230433. [Google Scholar] [CrossRef] [PubMed]
- Caniglia, R.; Fabbri, E.; Greco, C.; Galaverni, M.; Randi, E. Forensic DNA against Wildlife Poaching: Identification of a Serial Wolf Killing in Italy. Forensic Sci. Int. 2010, 4, 334–338. [Google Scholar] [CrossRef]
- Fabbri, E.; Velli, E.; D’Amico, F.; Galaverni, M.; Mastrogiuseppe, L.; Mattucci, F.; Caniglia, R. From Predation to Management: Monitoring Wolf Distribution and Understanding Depredation Patterns from Attacks on Livestock. Hystrix Ital. J. Mammal. 2018, 29, 101–110. [Google Scholar] [CrossRef]
- Mattucci, F.; Oliveira, R.; Bizzarri, L.; Vercillo, F.; Anile, S.; Ragni, B.; Lapini, L.; Sforzi, A.; Alves, P.C.; Lyons, L.A.; et al. Genetic Structure of Wildcat (Felis silvestris) Populations in Italy. Ecol. Evol. 2013, 3, 2443–2458. [Google Scholar] [CrossRef]
- Teerink, B.J. Hair of West-European Mammals: Atlas and Identification Key; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Taberlet, P. Reliable Genotyping of Samples with Very Low DNA Quantities Using PCR. Nucleic Acids Res. 1996, 24, 3189–3194. [Google Scholar] [CrossRef] [PubMed]
- Menotti-Raymond, M.; O’Brien, S.J. Evolutionary Conservation of Ten Microsatellite Loci in Four Species of Felidae. J. Hered. 1995, 86, 319–322. [Google Scholar] [CrossRef]
- MenottiRaymond, M.A.; David, V.A.; Obrien, S.J. Pet Cat Hair Implicates Murder Suspect. Nature 1997, 386, 774. [Google Scholar] [CrossRef]
- Valière, N.; Valière, N. Gimlet: A Computer Program for Analysing Genetic Individual Identification Data. Mol. Ecol. Notes 2002, 2, 377–379. [Google Scholar] [CrossRef]
- Miller, D.H.; Jensen, A.L.; Hammill, J.H. Density Dependent Matrix Model for Gray Wolf Population Projection. Ecol. Modell. 2002, 151, 271–278. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.K.; Pilgrim, K.L.; McKelvey, K.S.; Rivera, P.T.; Ruggiero, L.F. DNA Markers for Identifying Individual Snowshoe Hares Using Field-Collected Pellets. Northwest Sci. 2007, 81, 316–322. [Google Scholar] [CrossRef]
- Lukacs, P.M.; Eggert, L.S.; Burnham, K.P. Estimating Population Size from Multiple Detections with Non-Invasive Genetic Data. Wildl. Biol. Pract. 2007, 3, 83–92. [Google Scholar]
- Thuo, D.; Furlan, E.; Broekhuis, F.; Kamau, J.; MacDonald, K.; Gleeson, D.M. Food from Faeces: Evaluating the Efficacy of Scat DNA Metabarcoding in Dietary Analyses. PLoS ONE 2019, 15, e0228950. [Google Scholar] [CrossRef]
- Apostolico, F.; Vercillo, F.; La Porta, G.; Ragni, B. Long-Term Changes in Diet and Trophic Niche of the European Wildcat (Felis silvestris silvestris) in Italy. Mammal Res. 2016, 61, 109–119. [Google Scholar] [CrossRef]
- Stronen, A.V.; Aspi, J.; Caniglia, R.; Fabbri, E.; Galaverni, M.; Godinho, R.; Kvist, L.; Mattucci, F.; Nowak, C.; von Thaden, A.; et al. Wolf-Dog Admixture Highlights the Need for Methodological Standards and Multidisciplinary Cooperation for Effective Governance of Wild x Domestic Hybrids. Biol. Conserv. 2022, 266, 109467. [Google Scholar] [CrossRef]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G. Hybridization and Extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef]
- Canu, A.; Mattioli, L.; Santini, A.; Apollonio, M.; Scandura, M. “Video-Scats”: Combining Camera Trapping and Non-Invasive Genotyping to Assess Individual Identity and Hybrid Status in Gray Wolf. Wildlife Biol. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Anile, S.; Ragni, B.; Randi, E.; Mattucci, F.; Rovero, F. Wildcat Population Density on the Etna Volcano, Italy: A Comparison of Density Estimation Methods. J. Zool. 2014, 293, 252–261. [Google Scholar] [CrossRef]
- Newsome, T.M.; Boitani, L.; Chapron, G.; Ciucci, P.; Dickman, C.R.; Dellinger, J.A.; Lopez-Bao, J.V.; Peterson, R.O.; Shores, C.R.; Wirsing, A.J.; et al. Food Habits of the World’s Grey Wolves. Mamm. Rev. 2016, 46, 255–269. [Google Scholar] [CrossRef]
- Germain, E.; Ruette, S.; Poulle, M.L. Likeness between the Food Habits of European Wildcats, Domestic Cats and Their Hybrids in France. Mamm. Biol. 2009, 74, 412–417. [Google Scholar] [CrossRef]
- Scandura, M.; Iacolina, L.; Apollonio, M. Genetic Diversity In The European Wild Boar Sus scrofa: Phylogeography, Population Structure And Wild X Domestic Hybridization. Mamm. Rev. 2011, 41, 125–137. [Google Scholar] [CrossRef]
- Lorenzini, R.; Fanelli, R.; Tancredi, F.; Siclari, A.; Garofalo, L. Matching STR and SNP Genotyping to Discriminate between Wild Boar, Domestic Pigs and Their Recent Hybrids for Forensic Purposes. Sci. Rep. 2020, 10, 3188. [Google Scholar] [CrossRef]
- Dziech, A. Identification of Wolf-Dog Hybrids in Europe—An Overview of Genetic Studies. Front. Ecol. Evol. 2021, 9, 760160. [Google Scholar] [CrossRef]
- Harmoinen, J.; von Thaden, A.; Aspi, J.; Kvist, L.; Cocchiararo, B.; Jarausch, A.; Gazzola, A.; Sin, T.; Lohi, H.; Hytönen, M.K.; et al. Reliable Wolf-Dog Hybrid Detection in Europe Using a Reduced SNP Panel Developed for Non-Invasively Collected Samples. BMC Genom. 2021, 22, 473. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velli, E.; Mattucci, F.; Lazzeri, L.; Fabbri, E.; Pacini, G.; Belardi, I.; Mucci, N.; Caniglia, R. “Guess Who’s Coming to Dinner”: Molecular Tools to Reconstruct multilocus Genetic Profiles from Wild Canid Consumption Remains. Animals 2022, 12, 2428. https://doi.org/10.3390/ani12182428
Velli E, Mattucci F, Lazzeri L, Fabbri E, Pacini G, Belardi I, Mucci N, Caniglia R. “Guess Who’s Coming to Dinner”: Molecular Tools to Reconstruct multilocus Genetic Profiles from Wild Canid Consumption Remains. Animals. 2022; 12(18):2428. https://doi.org/10.3390/ani12182428
Chicago/Turabian StyleVelli, Edoardo, Federica Mattucci, Lorenzo Lazzeri, Elena Fabbri, Giada Pacini, Irene Belardi, Nadia Mucci, and Romolo Caniglia. 2022. "“Guess Who’s Coming to Dinner”: Molecular Tools to Reconstruct multilocus Genetic Profiles from Wild Canid Consumption Remains" Animals 12, no. 18: 2428. https://doi.org/10.3390/ani12182428






