The 16S rRNA Gene Sequencing of Gut Microbiota in Chickens Infected with Different Virulent Newcastle Disease Virus Strains
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Ethical Statement
2.3. Experiment Design
2.4. DNA Extraction and Library Construction
2.5. Bioinformatic Analysis
3. Results
3.1. Clinical Symptoms Post NDV Challenge
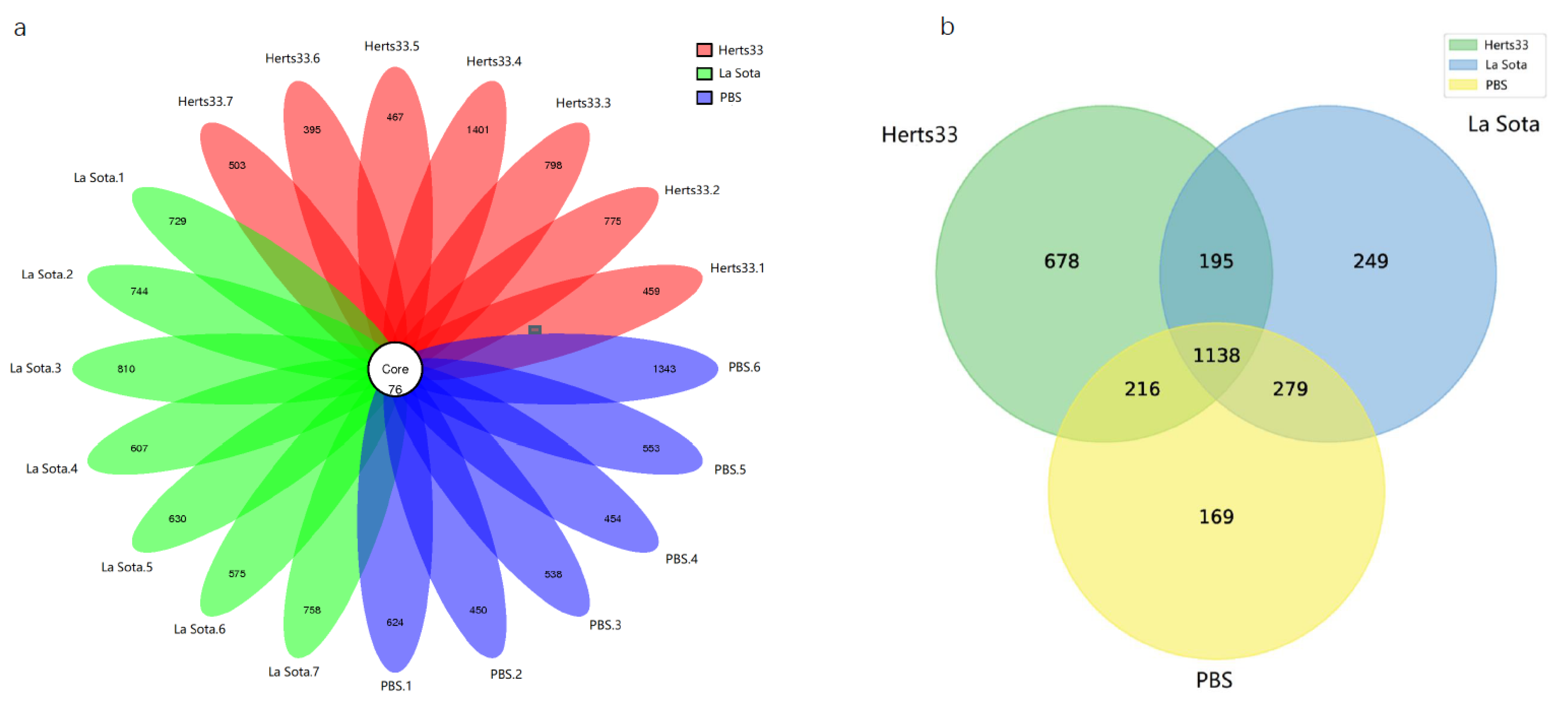
3.2. Sequencing Results Overview
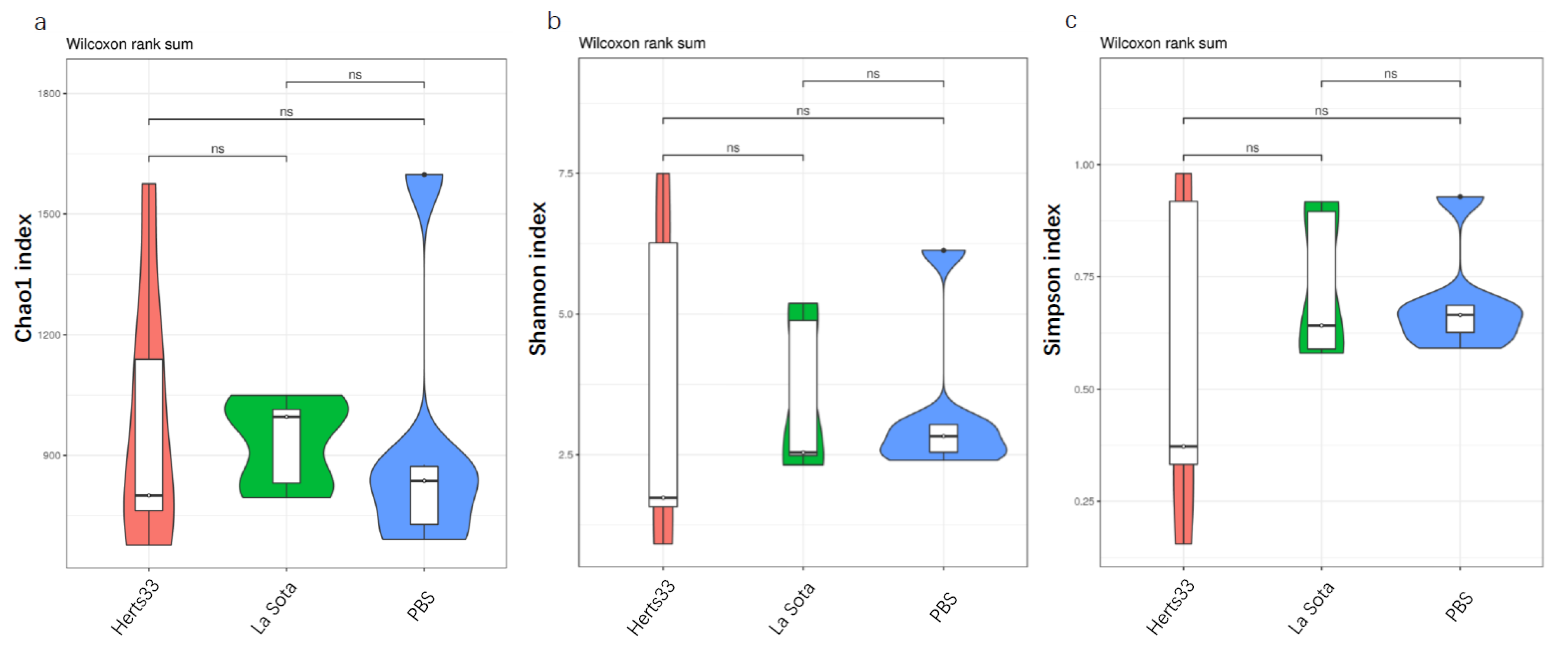
3.3. A Decrease in the Microbial Diversity in Gut Microbiota with NDV Infection
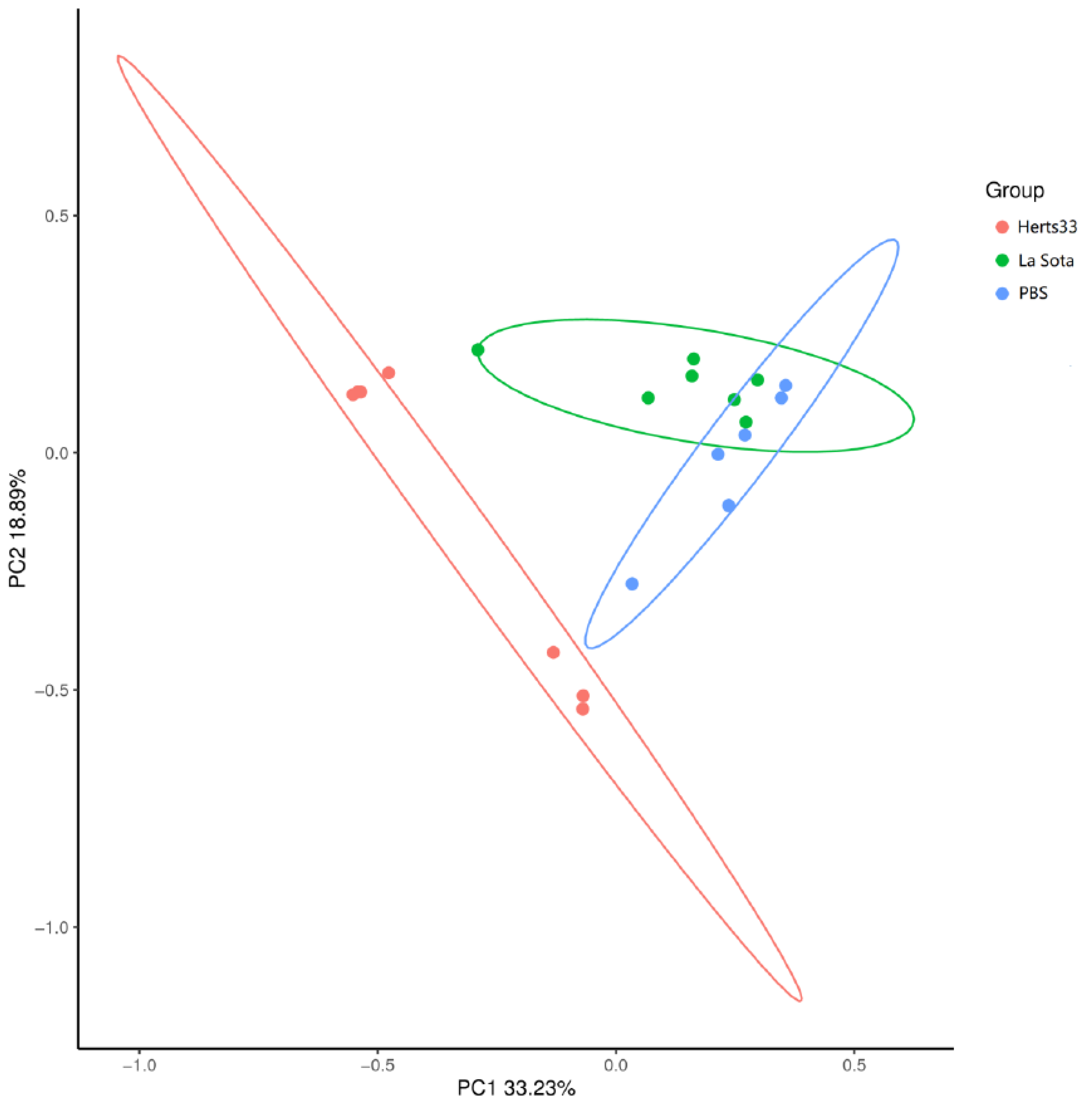
3.4. Gut Bacterial Beta-Diversity Analysis
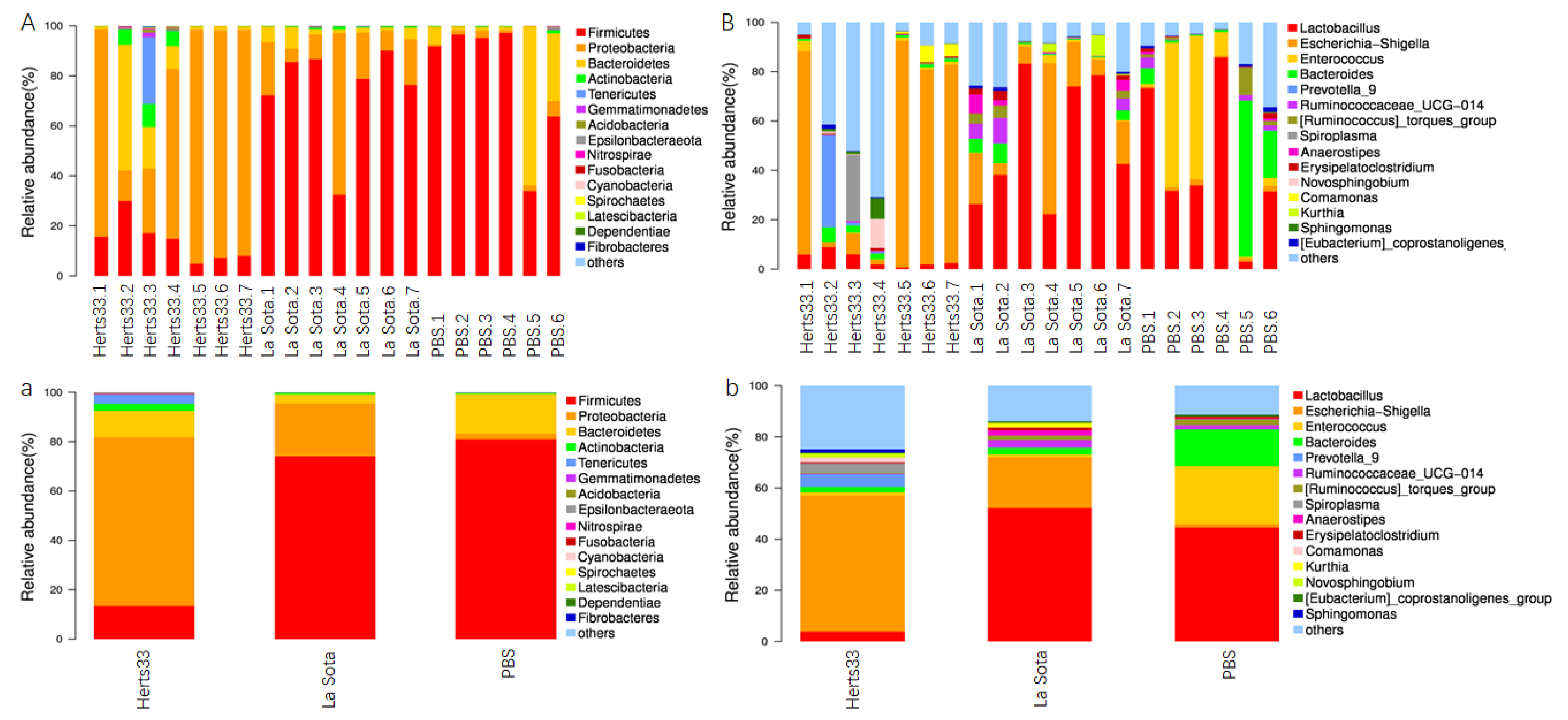
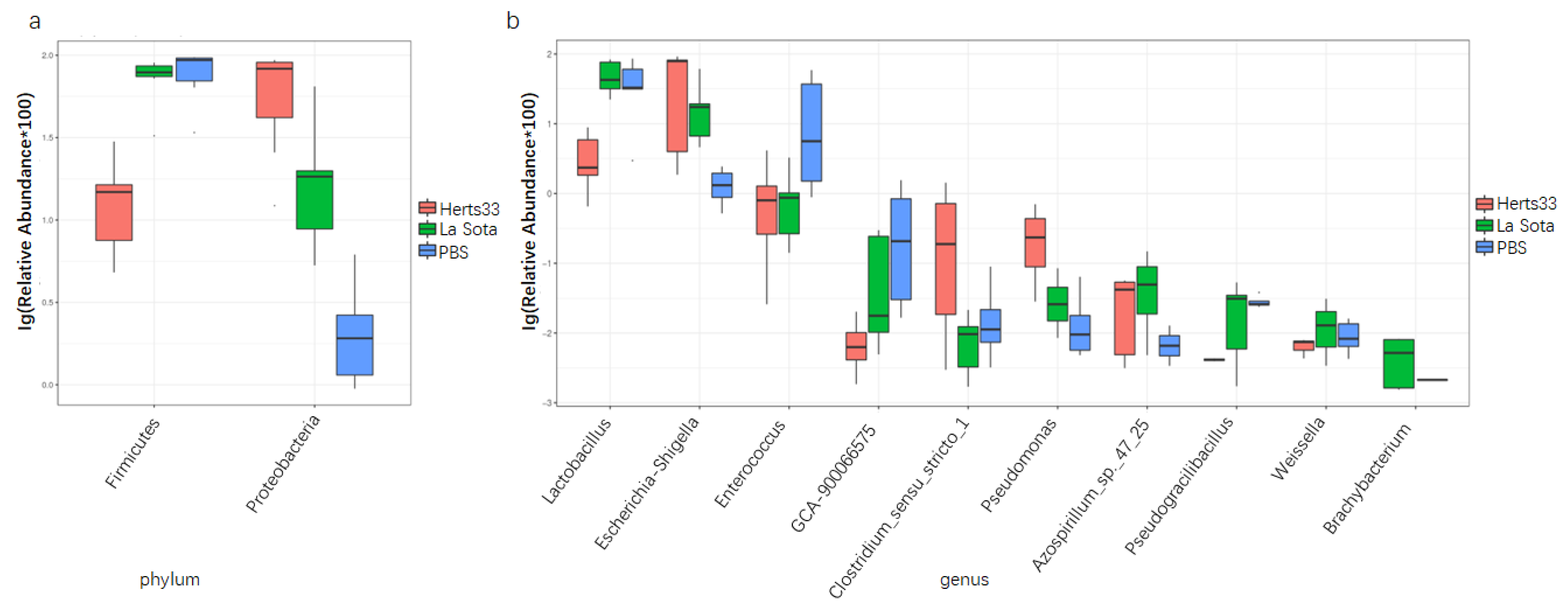
3.5. NDV Infection Alter the Gut Microbiome Composition in Chickens
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norman, J.M.; Handley, S.A.; Virgin, H.W. Kingdom-Agnostic Metagenomics and the Importance of Complete Characterization of Enteric Microbial Communities. Gastroenterology 2014, 146, 1459–1469. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; Conway, S.; Aminov, R. Commensal gut bacteria: Mechanisms of immune modulation. Trends Immunol. 2005, 26, 326–333. [Google Scholar] [CrossRef]
- Roto, S.M.; Kwon, Y.M.; Ricke, S.C. Applications of In Ovo Technique for the Optimal Development of the Gastrointestinal Tract and the Potential Influence on the Establishment of Its Microbiome in Poultry. Front. Vet. Sci. 2016, 3, 63. [Google Scholar] [CrossRef]
- Varmuzova, K.; Kubasova, T.; Davidova-Gerzova, L.; Sisak, F.; Havlickova, H.; Sebkova, A.; Faldynova, M.; Rychlik, I. Composition of Gut Microbiota Influences Resistance of Newly Hatched Chickens to Salmonella Enteritidis Infection. Front. Microbiol. 2016, 7, 957. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, I. Composition and Function of Chicken Gut Microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLoS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Chen, F.; Zuo, K.; Wu, C.; Yan, Y.; Chen, W.; Lin, W.; Xie, Q. Avian Influenza Virus Subtype H9N2 Affects Intestinal Microbiota, Barrier Structure Injury, and Inflammatory Intestinal Disease in the Chicken Ileum. Viruses 2018, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.-X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell–dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Shi, Y.; Liu, P.; Yang, Y.; Zhou, C.; Li, G.; Luo, J.; Zhang, C.; Cao, H.; Hu, G.; et al. 16S rRNA gene sequencing reveals an altered composition of the gut microbiota in chickens infected with a nephropathogenic infectious bronchitis virus. Sci. Rep. 2020, 10, 3556. [Google Scholar] [CrossRef]
- Perumbakkam, S.; Hunt, H.D.; Cheng, H.H. Marek’s disease virus influences the core gut microbiome of the chicken during the early and late phases of viral replication. FEMS Microbiol. Ecol. 2014, 90, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Perumbakkam, S.; Hunt, H.D.; Cheng, H.H. Differences in CD8αα and cecal microbiome community during proliferation and late cytolytic phases of Marek’s disease virus infection are associated with genetic resistance to Marek’s disease. FEMS Microbiol. Ecol. 2016, 92, fiw188. [Google Scholar] [CrossRef]
- Cui, N.; Huang, X.; Kong, Z.; Huang, Y.; Huang, Q.; Yang, S.; Zhang, L.; Xu, C.; Zhang, X.; Cui, Y. Newcastle Disease Virus Infection Interferes With the Formation of Intestinal Microflora in Newly Hatched Specific-Pathogen-Free Chicks. Front. Microbiol. 2018, 9, 900. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kubasová, T.; Rychlik, I.; Hoerr, F.J.; Rautenschlein, S. Infectious bursal disease virus infection leads to changes in the gut associated-lymphoid tissue and the microbiota composition. PLoS ONE 2018, 13, e0192066. [Google Scholar] [CrossRef] [PubMed]
- Dittoe, D.K.; Ricke, S.C.; Kiess, A.S. Organic Acids and Potential for Modifying the Avian Gastrointestinal Tract and Reducing Pathogens and Disease. Front. Vet. Sci. 2018, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.; Hernandez-Patlan, D.; Solis-Cruz, B.; Kwon, Y.M.; Arreguin, M.A.; Latorre, J.D.; Hernandez-Velasco, X.; Hargis, B.M.; Tellez-Isaias, G. Evaluation of the Antimicrobial and Anti-inflammatory Properties of Bacillus-DFM (Norum™) in Broiler Chickens Infected With Salmonella Enteritidis. Front. Vet. Sci. 2019, 6, 282. [Google Scholar] [CrossRef]
- Samal, S.K. Newcastle disease and related avian paramyxoviruses. In The Biology of Paramyxoviruses; Samal, S.K., Ed.; Caister Academic Press: Norfolk, UK, 2011; pp. 66–114. [Google Scholar]
- Alexander, D.J. Gordon Memorial Lecture. Newcastle disease. Br. Poult. Sci. 2001, 42, 5–22. [Google Scholar] [CrossRef]
- Reed, J.; Muench, H. A simple method of estimating fifty percent endpoint. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Chao, A.; Bunge, J. Estimating the Number of Species in a Stochastic Abundance Model. Biometrics 2002, 58, 531–539. [Google Scholar] [CrossRef]
- Hill, T.C.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.; Taylor, M.W. Characterizing the avian gut microbiota: Membership, driving influences, and potential function. Front. Microbiol. 2014, 5, 223. [Google Scholar] [CrossRef]
- Yegani, M.; Korver, D.R. Factors Affecting Intestinal Health in Poultry. Poult. Sci. 2008, 87, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J. The Avian Immune System. Disease of Poultry; Saif, Y.M., Ed.; Iowa State University Press: Ames, IA, USA, 2003; pp. 5–16. [Google Scholar]
- Pfeiffer, J.K.; Virgin, H.W. Viral immunity: Transkingdom control of viral infection and immunity in the mammalian intestine. Science 2016, 351, aad5872. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal Microbiota Promote Enteric Virus Replication and Systemic Pathogenesis. Science 2011, 334, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Morrison, M.; Yu, Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013, 92, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Lamendella, R.; Santo Domingo, J.W.; Ghosh, S.; Martinson, J.; Oerther, D.B. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011, 11, 103. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Holtrop, G.; Anderson, S.E.; Lobley, G.E.; Flint, H.J. Impact of pH on Lactate Formation and Utilization by Human Fecal Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 6526–6533. [Google Scholar] [CrossRef] [PubMed]
- Crhanova, M.; Karasova, D.; Juricova, H.; Matiasovicova, J.; Jahodarova, E.; Kubasova, T.; Seidlerova, Z.; Cizek, A.; Rychlik, I. Systematic Culturomics Shows that Half of Chicken Caecal Microbiota Members can be Grown in Vitro Except for Two Lineages of Clostridiales and a Single Lineage of Bacteroidetes. Microorganisms 2019, 7, 496. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Haghighi, H.R.; Gong, J.; Gyles, C.L.; Hayes, M.A.; Sanei, B.; Parvizi, P.; Gisavi, H.; Chambers, J.R.; Sharif, S. Modulation of Antibody-Mediated Immune Response by Probiotics in Chickens. Clin. Diagn. Lab. Immunol. 2005, 12, 1387–1392. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Huang, G.; Tang, X.; Bi, F.; Hao, Z.; Han, Z.; Suo, J.; Zhang, S.; Wang, S.; Duan, C.; Yu, Z.; et al. Eimeria tenella infection perturbs the chicken gut microbiota from the onset of oocyst shedding. Vet. Parasitol. 2018, 258, 30–37. [Google Scholar] [CrossRef]
- Yitbarek, A.; Alkie, T.; Taha-Abdelaziz, K.; Astill, J.; Rodriguez-Lecompte, J.C.; Parkinson, J.; Nagy, É.; Sharif, S. Gut microbiota modulates type I interferon and antibody-mediated immune responses in chickens infected with influenza virus subtype H9N2. Benef. Microbes 2018, 9, 417–427. [Google Scholar] [CrossRef]
- Yitbarek, A.; Taha-Abdelaziz, K.; Hodgins, D.C.; Read, L.; Nagy, É.; Weese, J.S.; Caswell, J.L.; Parkinson, J.; Sharif, S. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci. Rep. 2018, 8, 13189. [Google Scholar] [CrossRef]
- Nakamoto, N.; Amiya, T.; Aoki, R.; Taniki, N.; Koda, Y.; Miyamoto, K.; Teratani, T.; Suzuki, T.; Chiba, S.; Chu, P.-S.; et al. Commensal Lactobacillus Controls Immune Tolerance during Acute Liver Injury in Mice. Cell Rep. 2017, 21, 1215–1226. [Google Scholar] [CrossRef]
- Zenewicz, L.; Yin, X.; Wang, G.; Elinav, E.; Hao, L.; Zhao, L.; Flavell, R.A. IL-22 Deficiency Alters Colonic Microbiota To Be Transmissible and Colitogenic. J. Immunol. 2013, 190, 5306–5312. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Wang, T.-T.; Xu, M.; Evivie, S.E.; Luo, G.-W.; Liang, H.-Z.; Yu, S.-F.; Huo, G.-C. Effect of Lactobacillus Strains on Intestinal Microflora and Mucosa Immunity in Escherichia coli O157:H7-Induced Diarrhea in Mice. Curr. Microbiol. 2016, 73, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, Q.; Li, H.; Xu, C.; Cui, N.; Zhao, X. 16S rRNA genes Illumina sequencing revealed differential cecal microbiome in specific pathogen free chickens infected with different subgroup of avian leukosis viruses. Vet. Microbiol. 2017, 207, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Wang, X.; Wang, Q.; Li, H.; Wang, F.; Zhao, X. Effect of Dual Infection with Eimeria tenella and Subgroup J Avian Leukosis Virus on the Cecal Microbiome in Specific-Pathogen-Free Chicks. Front. Vet. Sci. 2017, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Du, E.; Gan, L.; Li, Z.; Wang, W.; Liu, D.; Guo, Y. In vitro antibacterial activity of thymol and carvacrol and their effects on broiler chickens challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 2015, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Kogut, M.H. Spatial and Temporal Changes in the Broiler Chicken Cecal and Fecal Microbiomes and Correlations of Bacterial Taxa with Cytokine Gene Expression. Front. Vet. Sci. 2016, 3, 11. [Google Scholar] [CrossRef]

| Samples | Reads | OTUs | Simpson | Chao1 | Observed Species | Shannon | Goods Coverage | PD Whole Tree |
|---|---|---|---|---|---|---|---|---|
| Herts33.1 | 67,293 | 535 | 0.315108 | 764.6402 | 447.8 | 1.40745 | 0.995131 | 21.40172 |
| Herts33.2 | 54,131 | 851 | 0.917592 | 1126.855 | 815.2 | 5.980166 | 0.995479 | 36.46119 |
| Herts33.3 | 57,483 | 874 | 0.918702 | 1151.576 | 820.5 | 6.545797 | 0.995068 | 37.76043 |
| Herts33.4 | 50,601 | 1477 | 0.980155 | 1575.387 | 1458.1 | 7.497044 | 0.995838 | 57.57711 |
| Herts33.5 | 69,273 | 543 | 0.155358 | 759.3362 | 449.8 | 0.914995 | 0.995384 | 21.97474 |
| Herts33.6 | 61,314 | 471 | 0.372028 | 677.053 | 416.3 | 1.730422 | 0.995881 | 20.37164 |
| Herts33.7 | 63,697 | 579 | 0.349542 | 799.8134 | 501.5 | 1.730623 | 0.99504 | 24.97967 |
| La Sota.1 | 58,778 | 805 | 0.913189 | 1006.878 | 746.1 | 5.120451 | 0.99454 | 30.27542 |
| La Sota.2 | 58,034 | 820 | 0.9171 | 1049.569 | 756.2 | 5.189811 | 0.994462 | 30.44099 |
| La Sota.3 | 61,316 | 886 | 0.584854 | 1022.178 | 815.2 | 2.510756 | 0.994423 | 35.28361 |
| La Sota.4 | 64,931 | 683 | 0.594488 | 824.6888 | 609.2 | 2.538315 | 0.995507 | 27.8734 |
| La Sota.5 | 62,199 | 706 | 0.580574 | 837.9335 | 634.3 | 2.316419 | 0.99486 | 27.89631 |
| La Sota.6 | 60,679 | 651 | 0.641917 | 794.4215 | 593.1 | 2.462201 | 0.99524 | 27.51244 |
| La Sota.7 | 57,813 | 834 | 0.877119 | 996.3602 | 774.2 | 4.652152 | 0.994438 | 31.2902 |
| PBS.1 | 61,439 | 700 | 0.619337 | 861.2555 | 630.9 | 3.045674 | 0.995023 | 26.75791 |
| PBS.2 | 59,118 | 526 | 0.647619 | 690.9104 | 476.7 | 2.510859 | 0.995636 | 21.37007 |
| PBS.3 | 60,339 | 614 | 0.683144 | 876.2117 | 544.2 | 2.643193 | 0.994499 | 24.31326 |
| PBS.4 | 62,201 | 530 | 0.687748 | 699.7192 | 463.5 | 2.40015 | 0.995292 | 22.87679 |
| PBS.5 | 55,910 | 629 | 0.591706 | 812.8232 | 585.7 | 3.014565 | 0.995425 | 24.2154 |
| PBS.6 | 46,932 | 1419 | 0.928459 | 1598.205 | 1411.9 | 6.129219 | 0.993234 | 51.03208 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, L.; Wang, W.; Ren, S.; Wang, J.; Wang, J.; Qu, Y.; Addoma Adam, F.E.; Li, Z.; Gao, X. The 16S rRNA Gene Sequencing of Gut Microbiota in Chickens Infected with Different Virulent Newcastle Disease Virus Strains. Animals 2022, 12, 2558. https://doi.org/10.3390/ani12192558
Tong L, Wang W, Ren S, Wang J, Wang J, Qu Y, Addoma Adam FE, Li Z, Gao X. The 16S rRNA Gene Sequencing of Gut Microbiota in Chickens Infected with Different Virulent Newcastle Disease Virus Strains. Animals. 2022; 12(19):2558. https://doi.org/10.3390/ani12192558
Chicago/Turabian StyleTong, Lina, Wen Wang, Shanhui Ren, Jianling Wang, Jie Wang, Yang Qu, Fathalrhman Eisa Addoma Adam, Zengkui Li, and Xiaolong Gao. 2022. "The 16S rRNA Gene Sequencing of Gut Microbiota in Chickens Infected with Different Virulent Newcastle Disease Virus Strains" Animals 12, no. 19: 2558. https://doi.org/10.3390/ani12192558
APA StyleTong, L., Wang, W., Ren, S., Wang, J., Wang, J., Qu, Y., Addoma Adam, F. E., Li, Z., & Gao, X. (2022). The 16S rRNA Gene Sequencing of Gut Microbiota in Chickens Infected with Different Virulent Newcastle Disease Virus Strains. Animals, 12(19), 2558. https://doi.org/10.3390/ani12192558







