Heteroscedastic Reaction Norm Models Improve the Assessment of Genotype by Environment Interaction for Growth, Reproductive, and Visual Score Traits in Nellore Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Dataset Description
2.2. Environment Descriptor
2.3. Reaction Norm Models (RNM)
2.4. Model Comparison
2.5. Environmental Sensitivity
3. Results
3.1. Reaction Norm Models
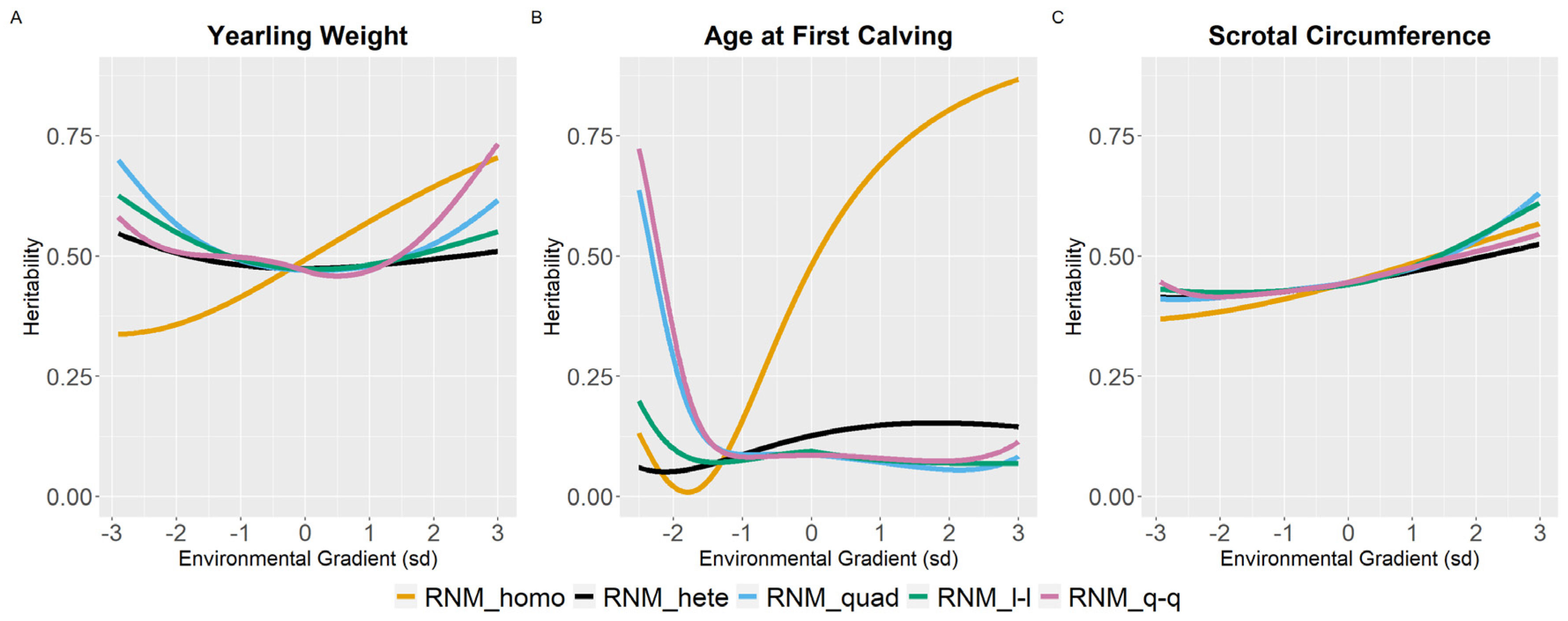
3.2. Heritability Estimates
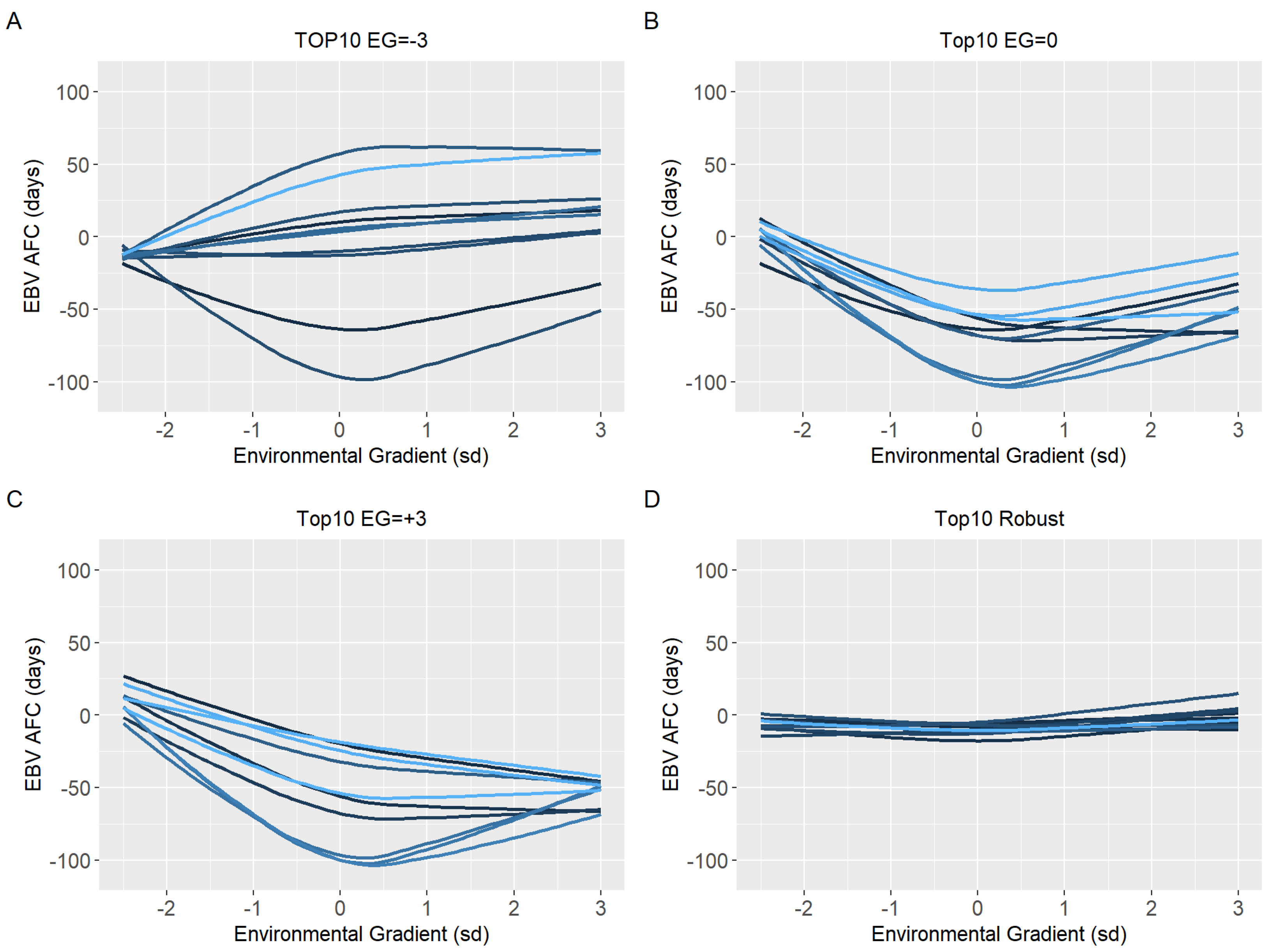
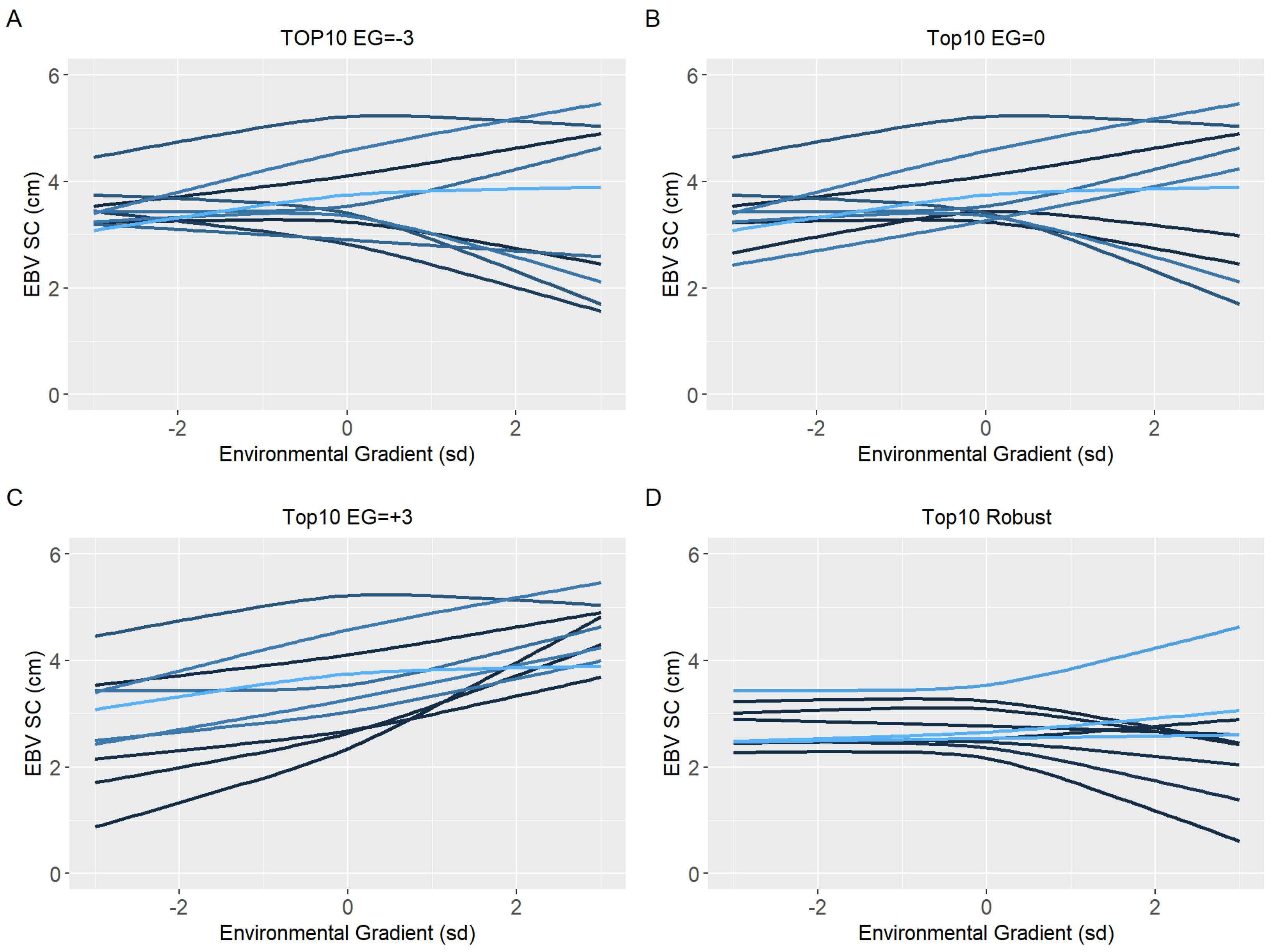
3.3. Environmental Sensitivity
4. Discussion
4.1. Reaction Norm Models
4.2. Heritability Estimates
4.3. Environmental Sensitivity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poppi, D.P.; Quigley, S.P.; da Silva, T.A.C.C.; McLennan, S.R. Challenges of beef cattle production from tropical pastures. Rev. Bras. Zootec. 2018, 47, e20160419. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; De Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Chiaia, H.L.J.; De Lemos, M.V.A.; Venturini, G.C.; Aboujaoude, C.; Berton, M.P.; Feitosa, F.B.; Carvalheiro, R.; Albuquerque, L.G.; de Oliveira, H.N.; Baldi, F. Genotype × environment interaction for age at first calving, scrotal circumference, and yearling weight in Nellore cattle using reaction norms in multitrait random regression models. J. Anim. Sci. 2015, 93, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- de Araujo Neto, F.R.; Pegolo, N.T.; Aspilcueta-borquis, R.R.; Pessoa, M.C.; Bonifácio, A.; Lobo, R.B.; de Oliveira, H.N. Study of the effect of genotype × environment interaction on age at first calving and production traits in Nellore cattle using multi-trait reaction norms and Bayesian inference. Anim. Sci. J. 2018, 89, 939–945. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Costa, L.S.; Garzón, N.A.M.; Passafaro, T.L.; Silva, D.O.; Abreu, L.R.A.; Verardo, L.L.; Bonafé, C.M.; Ventura, H.T. Unraveling the effect of body structure score on phenotypic plasticity for body weight at di ff erent ages in Guzerat cattle. Livest. Sci. 2019, 229, 98–104. [Google Scholar] [CrossRef]
- Ambrosini, D.P.; Henrique, C.; Malhado, M.; Filho, R.M.; Cardoso, F.F.; Luiz, P.; Carneiro, S. Genotype × environment interactions in reproductive traits of Nellore cattle in northeastern Brazil. Trop. Anim. Health Prod. 2016, 48, 1401–1407. [Google Scholar] [CrossRef]
- Mota, R.R.; Tempelman, R.J.; Lopes, P.S.; Aguilar, I.; Silva, F.F.; Cardoso, F.F. Genotype by environment interaction for tick resistance of Hereford and Braford beef cattle using reaction norm models. Genet. Sel. Evol. 2016, 48, 3. [Google Scholar] [CrossRef]
- Oliveira, D.P.; Lourenco, D.A.L.; Tsuruta, S.; Misztal, I.; Santos, D.J.A.; de Araújo Neto, F.R.; Aspilcueta-Borquis, R.R.; Baldi, F.; Carvalheiro, R.; de Camargo, G.M.F.; et al. Reaction norm for yearling weight in beef cattle using single-step genomic evaluation1. J. Anim. Sci. 2018, 96, 27–34. [Google Scholar] [CrossRef]
- Carvalheiro, R.; Costilla, R.; Neves, H.H.R.; Albuquerque, L.G.; Moore, S.; Hayes, B.J. Unraveling genetic sensitivity of beef cattle to environmental variation under tropical conditions. Genet. Sel. Evol. 2019, 51, 29. [Google Scholar] [CrossRef]
- Carvalho, C.V.D.; Costa, R.B.; de Camargo, G.M.F.; Bittencourt, T.C.C. Genotype × Environment Interaction for reproductive traits in brazilian Nellore breed cattle. Rev. Bras. Saude e Prod. Anim. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Silva, T.D.L.; Carneiro, P.L.S.; Ambrosini, D.P.; Lobo, R.B.; Martins Filho, R.; Malhado, C.H.M. Genotype-environment interaction in the genetic variability analysis of reproductive traits in Nellore cattle. Livest. Sci. 2019, 230, 103825. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Fernandes, G.A., Jr.; Herrera, A.C.; Scalez, D.C.B.; Espigolan, R.; Carvalheiro, R.; Baldi, F.; Albuquerque, L.G. Genomic reaction norm models exploiting genotype × environment interaction on sexual precocity indicator traits in Nellore cattle. Anim. Genet. 2020, 51, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L.R. Application of random regression models in animal breeding. Livest. Prod. Sci. 2004, 86, 35–45. [Google Scholar] [CrossRef]
- Rezende, M.P.G.; Malhado, C.H.M.; Biffani, S.; Carneiro, P.L.S.; Carrillo, J.A.; Bozzi, R. Genotype-environment interaction for age at first calving in Limousine and Charolais cattle raised in Italy, employing reaction norm model. Livest. Sci. 2020, 232, 103912. [Google Scholar] [CrossRef]
- Toghiani, S.; Hay, E.; Fragomeni, B.; Rekaya, R.; Roberts, A.J. Genotype by environment interaction in response to cold stress in a composite beef cattle breed. Animal 2020, 14, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.D.P.; Santana Júnior, M.L.; Schenkel, F.S.; Mercadante, M.E.Z.; Cyrillo, J.N.D.S.G.; de Paz, C.C.P. Different selection practices affect the environmental sensitivity of beef cattle. PLoS ONE 2021, 16, e0248186. [Google Scholar] [CrossRef]
- Negri, R.; Aguilar, I.; Feltes, G.L.; Cobuci, J.A. Selection for Test-Day Milk Yield and Thermotolerance in Brazilian Holstein Cattle. Animals 2021, 11, 128. [Google Scholar] [CrossRef]
- Shi, R.; Brito, L.F.; Liu, A.; Luo, H.; Chen, Z.; Liu, L.; Guo, G.; Mulder, H.; Ducro, B.; van der Linden, A.; et al. Genotype-by-environment interaction in Holstein heifer fertility traits using single-step genomic reaction norm models. BMC Genom. 2021, 22, 193. [Google Scholar] [CrossRef]
- Strandberg, E.; Kolmodin, R.; Madsen, P.; Jensen, J.; Jorjani, H. Genotype by Environment Interaction in Nordic Dairy Cattle Studied by Use of Reaction Norms. Interbull Bull. 2000, 25, 41. [Google Scholar]
- R Core Team; A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Kolmodin, R.; Strandberg, E.; Madsen, P.; Jensen, J.; Jorjani, H. Genotype by Environment Interaction in Nordic Dairy Cattle Studied Using Reaction Norms. Acta Agric. Scand. 2002, 52, 11–24. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; pp. 2001–2002.
- Foulley, J.L.; Quaas, R.L. Heterogeneous variances in Gaussian linear mixed models. Genet. Sel. Evol. 1995, 27, 211. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Multimodel Inference Understanding AIC and BIC in Model Selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Sanata, M.L., Jr.; Bignardi, A.B.; Pereira, R.J. Random regression models to account for the effect of genotype by environment interaction due to heat stress on the milk yield of Holstein cows under tropical conditions. J. Appl. Genet. 2016, 57, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Daetwyler, H.D.; Goddard, M.E. Models for Genome × Environment interaction: Examples in livestock. Crop Sci. 2016, 56, 2251–2259. [Google Scholar] [CrossRef]
- Ambrosini, D.P.; Malhado, C.H.M.; Braccini Neto, J.; Martins Filho, R.; Affonso, P.R.A.D.M.; Luiz, P.S.C. Reaction norms of direct and maternal effects for weight at 205 days in Polled Nellore cattle in north-eastern Brazil. Arch. Tierz. 2014, 57, 1–11. [Google Scholar] [CrossRef]
- Calus, M.P.L.; Groen, A.F.; De Jong, G. Genotype × Environment Interaction for Protein Yield in Dutch Dairy Cattle as Quantified by Different Models. J. Dairy Sci. 2002, 85, 3115–3123. [Google Scholar] [CrossRef]
- Cardoso, L.L.; Neto, J.B.; Cardoso, F.F.; Araújo, J.; Biassus, I.D.O.; Otávio, J.; Barcellos, J. Hierarchical Bayesian models for genotype × environment estimates in post-weaning gain of Hereford bovine via reaction norms. Rev. Bras. Zootec. 2011, 40, 294–300. [Google Scholar] [CrossRef]
- Streit, M.; Reinhardt, F.; Thaller, G.; Bennewitz, J. Reaction norms and genotype-by-environment interaction in the German Holstein dairy cattle. J. Anim. Breed. Genet. 2012, 129, 380–389. [Google Scholar] [CrossRef]
- Meyer, K. Estimates of genetic covariance functions for growth of Angus cattle. J. Anim. Breed. Genet. 2005, 122, 73–85. [Google Scholar] [CrossRef]
- Ambrosini, D.P.; Carneiro, P.L.S.; Bracicini Neto, J.; Martins Filho, R.; Amaral, R.D.S.; Cardoso, F.F.; Malhado, C.H.M. Reaction norms models in the adjusted weight at 550 days of age for Polled Nellore cattle in Northeast Brazil. Rev. Bras. Zootec. 2014, 43, 351–357. [Google Scholar] [CrossRef]
- Ribeiro, S.; Eler, J.P.; Pedrosa, V.B.; Rosa, G.J.M.; Ferraz, J.B.S.; Balieiro, J.C.C. Genotype by environment interaction for yearling weight in Nellore cattle applying reaction norms models. Anim. Prod. Sci. 2017, 58, 1996–2002. [Google Scholar] [CrossRef]
- Lemos, M.V.A.; Chiaia, H.L.J.; Berton, M.P.; Feitosa, F.L.B.; Aboujaoude, C.; Venturini, G.C.; Oliveira, H.N.; Albuquerque, L.G.; Baldi, F. Reaction norms for the study of genotype- environment interaction for growth and indicator traits of sexual precocity in Nellore cattle. Genet. Mol. Res. 2015, 14, 7151–7162. [Google Scholar] [CrossRef]
- Cardoso, F.F.; Tempelman, R.J. Linear reaction norm models for genetic merit prediction of Angus cattle under genotype by environment interaction. J. Anim. Sci. 2012, 90, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Macneil, M.D.; Cardoso, F.F.; Hay, E. Genotype by environment interaction effects in genetic evaluation of preweaning gain for Line 1 Hereford cattle from Miles City, Montana 1. J. Anim. Sci. 2017, 95, 3833–3838. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K. Random regression analyses using B-splines to model growth of Australian Angus cattle. Genet. Sel. Evol. 2005, 37, 473–500. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I. Properties of random regression models using linear splines. J. Anim. Breed. Genet. 2006, 123, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Schenkel, F.S.; Brito, L.F. Unravelling Biological Biotypes for Growth, Visual Score and Reproductive Traits in Nellore Cattle via Principal Component Analysis. Livest. Sci. 2018, 217, 37–43. [Google Scholar] [CrossRef]
- Sigurdsson, A.; Banos, G.; Philipsson, J. Estimation of Genetic (Co)variance Components for International Evaluation of Dairy Bulls. Acta Agric. Scand. Sect. A—Anim. Sci. 1996, 46, 129–136. [Google Scholar] [CrossRef]
- Hammami, H.; Rekik, B.; Soyeurt, H.; Bastin, C.; Bay, E.; Stoll, J.; Gengler, N. Accessing genotype by environment interaction using within- and across-country test-day random regression sire models. J. Anim. Breed. Genet. 2009, 126, 366–377. [Google Scholar] [CrossRef]
- Santana, M.L.; Eler, J.P.; Cardoso, F.F.; Albuquerque, L.G.; Ferraz, J.B.S. Phenotypic plasticity of composite beef cattle performance using reaction norms model with unknown covariate. Animal 2013, 7, 202–210. [Google Scholar] [CrossRef]
- Su, G.; Madsen, P.; Lund, M.S.; Sorensen, D.; Korsgaard, I.R.; Jensen, J. Bayesian analysis of the linear reaction norm model with unknown covariates Bayesian analysis of the linear reaction norm model with unknown covariates 1. J. Anim. Sci. 2006, 84, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kargo, M.; Liu, A.; Thomasen, J.R.; Pan, Y.; Su, G. Genotype-by-environment interaction of fertility traits in Danish Holstein cattle using a single-step genomic reaction norm model. Heredity 2019, 123, 202–214. [Google Scholar] [CrossRef] [PubMed]





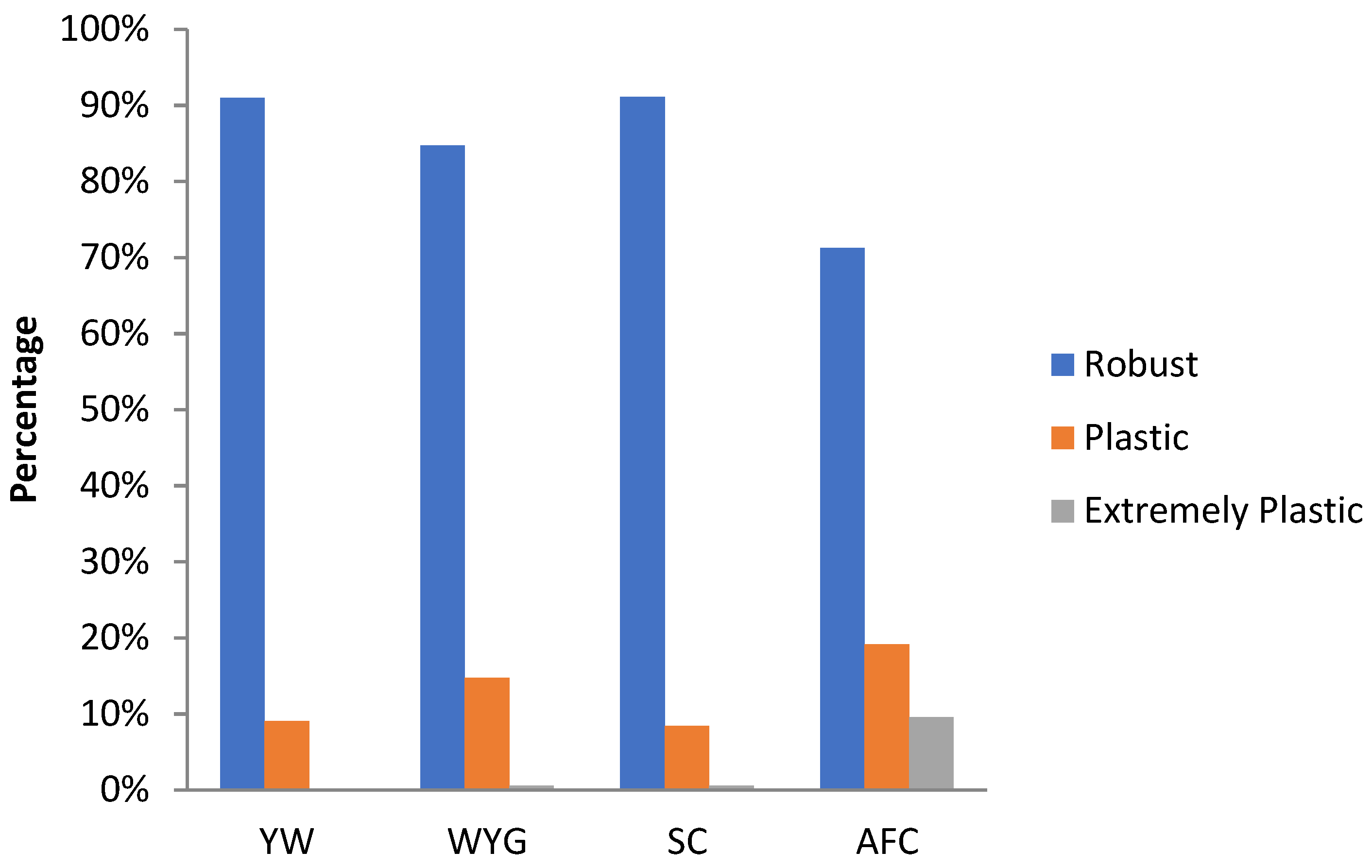
| Traits 1 | N | Female | Male | Min | Mean | Max | sd | CG |
|---|---|---|---|---|---|---|---|---|
| BWG | 553,381 | 276,994 | 276,387 | 70 | 157.2 | 278 | 32.95 | 11,657 |
| WC | 553,381 | 276,994 | 276,387 | 1 | 3 a | 5 | - | 11,657 |
| WP | 553,381 | 276,994 | 276,387 | 1 | 3 a | 5 | - | 11,657 |
| WM | 553,381 | 276,994 | 276,387 | 1 | 3 a | 5 | - | 11,657 |
| YW | 457,118 | 233,320 | 223,798 | 150 | 293 | 500 | 51.34 | 10,583 |
| WYG | 442,086 | 223,468 | 218,618 | 30 | 104.3 | 250 | 37.29 | 10,306 |
| YC | 529,673 | 270,252 | 259,421 | 1 | 3 a | 5 | - | 7246 |
| YP | 529,673 | 270,252 | 259,421 | 1 | 3 a | 5 | - | 7246 |
| YM | 529,673 | 270,252 | 259,421 | 1 | 3 a | 5 | - | 7246 |
| SC | 444,675 | - | 444,675 | 15 | 26.7 | 45 | 3.83 | 10,099 |
| AFC | 140,162 | 140,162 | - | 544 | 1012 | 1220 | 132 | 3897 |
| Variable | Trait 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BWG | WC | WP | WM | YW | WYG | YC | YP | YM | SC | AFC | |
| Birth year | X | X | X | X | X | X | X | X | X | X | X |
| Birth season | X | X | X | X | X | X | X | X | X | X | X |
| Sex | X | X | X | X | X | X | X | X | X | ||
| Farm at birth | X | X | X | X | X | X | X | X | X | ||
| Farm at weaning | X | X | X | X | X | X | X | X | |||
| Weaning management group | X | X | X | X | X | X | X | X | |||
| Yearling management group | X | X | X | X | X | X | X | ||||
| Farm at yearling | X | X | X | X | X | X | X | ||||
| Trait 1 | Model 2 | Coefficient 3 | b0 | b1 | b2 | b3 | σ2m | σ2e 4 | Np 5 | AIC | AICw |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BWG | RNM_homo | b0 (int) | 55.67 | −1.51 | 204.08 | 5 | 3,173,163.90 | 0.00 | |||
| b1 (slp) | −0.29 | 0.47 | |||||||||
| maternal | 113.94 | ||||||||||
| RNM_hete | b0 (int) | 55.07 | −0.18 | 5.32 | 6 | 3,173,157.20 | 0.00 | ||||
| b1 (slp) | −0.04 | 0.35 | −0.01 | ||||||||
| maternal | 114.05 | ||||||||||
| RNM_quad | b0 (int) | 55.88 | −0.94 | −0.83 | 5.31 | 10 | 3173156.30 | 0.00 | |||
| b1 (slp) | −0.11 | 1.38 | 0.45 | −0.02 | |||||||
| b2 (qdr) | −0.15 | 0.51 | 0.57 | 0.01 | |||||||
| maternal | 114.06 | ||||||||||
| RNM_l-l | b0 (int) | 56.98 | 0.84 | −3.75 | 5.30 | 10 | 3,173,160.00 | 0.00 | |||
| b1 (slp1) | 0.09 | 1.61 | −1.58 | −0.04 | |||||||
| b2 (slp2) | −0.21 | −0.53 | 5.54 | 0.04 | |||||||
| maternal | 114.06 | ||||||||||
| RNM_q-q | b0 (int) | 55.77 | −1.95 | −1.59 | 1.88 | 5.31 | 15 | 3,173,130.20 | 1.00 | ||
| b1 (slp1) | −0.14 | 3.37 | 1.48 | −2.90 | −0.06 | ||||||
| b2 (qdr1) | −0.19 | 0.73 | 1.23 | −1.69 | −0.02 | ||||||
| b3 (qrd2) | 0.13 | −0.83 | −0.80 | 3.66 | 0.06 | ||||||
| maternal | 114.00 | ||||||||||
| WC | RNM_homo | b0 (int) | 0.1428 | 0.0044 | 0.7004 | 5 | 33,076.51 | 0.06 | |||
| b1 (slp) | 0.62 | 0.0004 | |||||||||
| maternal | 0.2300 | ||||||||||
| RNM_hete | b0 (int) | 0.1426 | 0.0013 | −0.3559 | 6 | 33,070.84 | 0.94 | ||||
| b1 (slp) | 0.30 | 0.0001 | 0.0107 | ||||||||
| maternal | 0.2301 | ||||||||||
| RNM_quad | b0 (int) | 0.1523 | 0.0031 | −0.0078 | −0.3649 | 10 | 33,257.81 | 0.00 | |||
| b1 (slp) | 0.09 | 0.0077 | 0.0007 | 0.0055 | |||||||
| b2 (qdr) | −0.38 | 0.15 | 0.0027 | −0.0018 | |||||||
| maternal | 0.2304 | ||||||||||
| RNM_l-l | b0 (int) | 0.1510 | 0.0121 | −0.0177 | −0.3657 | 10 | 33,159.66 | 0.00 | |||
| b1 (slp1) | 0.33 | 0.0091 | −0.0123 | 0.0001 | |||||||
| b2 (slp2) | −0.28 | −0.78 | 0.0273 | 0.0117 | |||||||
| maternal | 0.2302 | ||||||||||
| RNM_q-q | b0 (int) | 0.1470 | 0.0081 | −0.0011 | −0.0059 | −0.3640 | 15 | 33,210.89 | 0.00 | ||
| b1 (slp1) | 0.13 | 0.0253 | 0.0108 | −0.0237 | 0.0029 | ||||||
| b2 (qdr1) | −0.04 | 0.86 | 0.0062 | −0.0113 | −0.0007 | ||||||
| b3 (qrd2) | −0.10 | −0.93 | −0.89 | 0.0259 | 0.0022 | ||||||
| maternal | 0.2303 | ||||||||||
| WP | RNM_homo | b0 (int) | 0.2349 | 0.0026 | 0.7900 | 5 | 104,255.53 | 0.55 | |||
| b1 (slp) | 0.33 | 0.0003 | |||||||||
| maternal | 0.2056 | ||||||||||
| RNM_hete | b0 (int) | 0.2349 | 0.0014 | −0.2359 | 6 | 104,255.91 | 0.45 | ||||
| b1 (slp) | 0.17 | 0.0003 | 0.0049 | ||||||||
| maternal | 0.2056 | ||||||||||
| RNM_quad | b0 (int) | 0.2426 | 0.0006 | −0.0076 | −0.2381 | 10 | 104,425.75 | 0.00 | |||
| b1 (slp) | 0.01 | 0.0072 | 0.0009 | 0.0030 | |||||||
| b2 (qdr) | −0.29 | 0.19 | 0.0029 | −0.0066 | |||||||
| maternal | 0.2065 | ||||||||||
| RNM_l-l | b0 (int) | 0.2431 | 0.0113 | −0.0199 | −0.2358 | 10 | 104,334.27 | 0.00 | |||
| b1 (slp1) | 0.23 | 0.0097 | −0.0148 | 0.0072 | |||||||
| b2 (slp2) | −0.22 | −0.82 | 0.0337 | −0.0118 | |||||||
| maternal | 0.2061 | ||||||||||
| RNM_q-q | b0 (int) | 0.2391 | −0.0006 | −0.0044 | 0.0018 | −0.2380 | 15 | 104,375.02 | 0.00 | ||
| b1 (slp1) | −0.01 | 0.0199 | 0.0092 | −0.0192 | 0.0023 | ||||||
| b2 (qdr1) | −0.12 | 0.84 | 0.0060 | −0.0102 | −0.0063 | ||||||
| b3 (qrd2) | 0.02 | −0.92 | −0.89 | 0.0222 | 0.0037 | ||||||
| maternal | 0.2059 | ||||||||||
| WM | RNM_homo | b0 (int) | 0.2078 | 0.0017 | 0.8269 | 5 | 128,693.45 | 0.73 | |||
| b1 (slp) | 0.12 | 0.0009 | |||||||||
| maternal | 0.2506 | ||||||||||
| RNM_hete | b0 (int) | 0.2078 | 0.0013 | -0.1902 | 6 | 128,695.39 | 0.27 | ||||
| b1 (slp) | 0.10 | 0.0009 | 0.0011 | ||||||||
| maternal | 0.2506 | ||||||||||
| RNM_quad | b0 (int) | 0.2160 | −0.0002 | −0.0073 | −0.1937 | 10 | 128,882.36 | 0.00 | |||
| b1 (slp) | −0.01 | 0.0074 | 0.0006 | 0.0023 | |||||||
| b2 (qdr) | −0.29 | 0.13 | 0.0029 | −0.0051 | |||||||
| maternal | 0.2512 | ||||||||||
| RNM_l-l | b0 (int) | 0.2160 | 0.0096 | −0.0180 | −0.1945 | 10 | 128,781.97 | 0.00 | |||
| b1 (slp1) | 0.20 | 0.0103 | −0.0150 | 0.0015 | |||||||
| b2 (slp2) | −0.22 | −0.83 | 0.0320 | 0.0005 | |||||||
| maternal | 0.2508 | ||||||||||
| RNM_q-q | b0 (int) | 0.2119 | −0.0015 | −0.0054 | 0.0030 | −0.1935 | 15 | 128,842.18 | 0.00 | ||
| b1 (slp1) | −0.02 | 0.0216 | 0.0099 | −0.0208 | 0.0103 | ||||||
| b2 (qdr1) | −0.15 | 0.84 | 0.0065 | −0.0110 | 0.0035 | ||||||
| b3 (qrd2) | 0.04 | −0.92 | −0.89 | 0.0238 | −0.0119 | ||||||
| maternal | 0.2511 |
| Traits 1 | Model 2 | Coefficient 3 | b0 | b1 | b2 | b3 | σ2e 4 | Np 5 | AIC | AICw |
|---|---|---|---|---|---|---|---|---|---|---|
| YW | RNM_homo | b0 (int) | 350.81 | 56.63 | 361.68 | 4 | 4,164,832 | 0.00 | ||
| b1 (slp) | 0.69 | 19.29 | ||||||||
| RNM_hete | b0 (int) | 336.55 | 23.99 | 5.92 | 5 | 4,164,587 | 0.00 | |||
| b1 (slp) | 0.38 | 12.14 | 0.14 | |||||||
| RNM_quad | b0 (int) | 336.43 | 22.33 | −1.68 | 5.94 | 9 | 4,164,537 | 0.00 | ||
| b1 (slp) | 0.31 | 15.23 | 0.03 | 0.15 | ||||||
| b2 (qdr) | −0.06 | 0.01 | 1.98 | −0.03 | ||||||
| RNM_l-l | b0 (int) | 343.00 | 30.15 | −16.51 | 5.95 | 9 | 4,164,520 | 1.00 | ||
| b1 (slp1) | 0.34 | 23.10 | −13.08 | 0.19 | ||||||
| b2 (slp2) | −0.17 | −0.52 | 26.99 | −0.09 | ||||||
| RNM_q-q | b0 (int) | 335.44 | 8.72 | −10.35 | 18.07 | 5.94 | 14 | 4,164,531 | 0.00 | |
| b1 (slp1) | 0.12 | 17.05 | 4.76 | −6.06 | 0.23 | |||||
| b2 (qdr1) | −0.25 | 0.52 | 4.97 | −7.02 | 0.04 | |||||
| b3 (qrd2) | 0.27 | −0.41 | −0.87 | 13.10 | −0.13 | |||||
| WYG | RNM_homo | b0 (int) | 113.46 | 35.97 | 261.28 | 4 | 3,780,797 | 0.00 | ||
| b1 (slp) | 0.89 | 14.49 | ||||||||
| RNM_hete | b0 (int) | 92.02 | 13.28 | 5.64 | 5 | 3,780,155 | 0.00 | |||
| b1 (slp) | 0.59 | 5.49 | 0.15 | |||||||
| RNM_quad | b0 (int) | 98.20 | 15.19 | −5.09 | 5.65 | 9 | 3,779,938 | 0.00 | ||
| b1 (slp) | 0.47 | 10.41 | −1.80 | 0.17 | ||||||
| b2 (qdr) | −0.40 | −0.44 | 1.63 | −0.03 | ||||||
| RNM_l-l | b0 (int) | 103.55 | 26.12 | −20.92 | 5.66 | 9 | 3,779,976 | 0.00 | ||
| b1 (slp1) | 0.56 | 20.68 | −15.95 | 0.20 | ||||||
| b2 (slp2) | −0.46 | −0.78 | 20.06 | −0.09 | ||||||
| RNM_q-q | b0 (int) | 97.78 | 8.25 | −7.79 | 6.07 | 5.64 | 14 | 3,779,882 | 1.00 | |
| b1 (slp1) | 0.21 | 15.09 | 8.03 | −9.56 | 0.22 | |||||
| b2 (qdr1) | −0.27 | 0.72 | 8.31 | −10.29 | 0.03 | |||||
| b3 (qrd2) | 0.17 | −0.68 | −0.98 | 13.24 | −0.09 | |||||
| AFC | RNM_homo | b0 (int) | 3391.50 | 1873.80 | 3668.90 | 4 | 1,597,302 | 0.00 | ||
| b1 (slp) | 1.00 | 1045.10 | ||||||||
| RNM_hete | b0 (int) | 828.06 | 309.89 | 8.65 | 5 | 1,593,825 | 0.00 | |||
| b1 (slp) | 0.93 | 133.86 | 0.46 | |||||||
| RNM_quad | b0 (int) | 651.88 | 150.00 | −107.08 | 8.84 | 9 | 1,592,165 | 0.00 | ||
| b1 (slp) | 0.60 | 94.51 | −31.44 | 0.56 | ||||||
| b2 (qdr) | −0.82 | −0.64 | 25.86 | −0.15 | ||||||
| RNM_l-l | b0 (int) | 746.31 | 331.13 | −354.60 | 8.88 | 9 | 1,592,410 | 0.00 | ||
| b1 (slp1) | 0.89 | 187.18 | −171.16 | 0.77 | ||||||
| b2 (slp2) | −0.89 | −0.86 | 210.83 | −0.52 | ||||||
| RNM_q-q | b0 (int) | 631.40 | 178.12 | −61.62 | −46.08 | 8.82 | 14 | 1,592,007 | 1.00 | |
| b1 (slp1) | 0.62 | 131.68 | 26.11 | −59.51 | 0.54 | |||||
| b2 (qdr1) | −0.30 | 0.28 | 67.02 | −64.41 | −0.17 | |||||
| b3 (qrd2) | −0.20 | −0.56 | −0.85 | 86.62 | 0.02 | |||||
| SC | RNM_homo | b0 (int) | 3.03 | 0.23 | 3.78 | 4 | 2,059,717.9 | 0.00 | ||
| b1 (slp) | 0.53 | 0.06 | ||||||||
| RNM_hete | b0 (int) | 3.02 | 0.17 | 1.33 | 5 | 2,059,691.4 | 0.00 | |||
| b1 (slp) | 0.43 | 0.05 | 0.03 | |||||||
| RNM_quad | b0 (int) | 3.06 | 0.15 | −0.04 | 1.34 | 9 | 2,059,612.4 | 0.00 | ||
| b1 (slp) | 0.26 | 0.11 | 0.02 | 0.02 | ||||||
| b2 (qdr) | −0.28 | 0.56 | 0.01 | −0.02 | ||||||
| RNM_l-l | b0 (int) | 3.07 | 0.20 | −0.13 | 1.36 | 9 | 2,059,618.7 | 0.00 | ||
| b1 (slp1) | 0.46 | 0.06 | −0.01 | 0.07 | ||||||
| b2 (slp2) | −0.18 | −0.07 | 0.18 | −0.10 | ||||||
| RNM_q-q | b0 (int) | 3.05 | 0.14 | −0.06 | 0.04 | 1.34 | 14 | 2,059,596.1 | 1.00 | |
| b1 (slp1) | 0.18 | 0.20 | 0.06 | −0.08 | −0.02 | |||||
| b2 (qdr1) | −0.20 | 0.72 | 0.03 | −0.04 | −0.04 | |||||
| b3 (qrd2) | 0.09 | −0.78 | −0.90 | 0.06 | 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho Filho, I.; Silva, D.A.; Teixeira, C.S.; Silva, T.L.; Mota, L.F.M.; Albuquerque, L.G.; Carvalheiro, R. Heteroscedastic Reaction Norm Models Improve the Assessment of Genotype by Environment Interaction for Growth, Reproductive, and Visual Score Traits in Nellore Cattle. Animals 2022, 12, 2613. https://doi.org/10.3390/ani12192613
Carvalho Filho I, Silva DA, Teixeira CS, Silva TL, Mota LFM, Albuquerque LG, Carvalheiro R. Heteroscedastic Reaction Norm Models Improve the Assessment of Genotype by Environment Interaction for Growth, Reproductive, and Visual Score Traits in Nellore Cattle. Animals. 2022; 12(19):2613. https://doi.org/10.3390/ani12192613
Chicago/Turabian StyleCarvalho Filho, Ivan, Delvan A. Silva, Caio S. Teixeira, Thales L. Silva, Lucio F. M. Mota, Lucia G. Albuquerque, and Roberto Carvalheiro. 2022. "Heteroscedastic Reaction Norm Models Improve the Assessment of Genotype by Environment Interaction for Growth, Reproductive, and Visual Score Traits in Nellore Cattle" Animals 12, no. 19: 2613. https://doi.org/10.3390/ani12192613







