Syringohydromyelia in Dogs: The Genomic Component Underlying a Complex Neurological Disease
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Whole-Genome Resequencing
2.3. Genome Wide-Association Studies (GWAS)
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rusbridge, C.; Greitz, D.; Iskandar, B.J. Syringomyelia: Current Concepts in Pathogenesis, Diagnosis, and Treatment. J. Veter. Intern. Med. 2006, 20, 469–479. [Google Scholar] [CrossRef]
- Vandertop, W.P. Syringomyelia. Neuropediatrics 2013, 45, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.E.; Knowler, S.P.; Rusbridge, C.; Noorman, E.; Jeffery, N.D. Prevalence of asymptomatic syringomyelia in Cavalier King Charles spaniels. Vet. Rec. 2011, 168, 667. [Google Scholar] [CrossRef] [PubMed]
- Rusbridge, C.; Knowler, S.P.; Pieterse, L.; McFadyen, A.K. Chiari-like malformation in the Griffon Bruxellois. J. Small Anim. Pract. 2009, 50, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Kiviranta, A.; Rusbridge, C.; Laitinen-Vapaavuori, O.; Hielm-Björkman, A.; Lappalainen, A.; Knowler, S.P.; Jokinen, T. Syringomyelia and Craniocervical Junction Abnormalities in Chihuahuas. J. Vet. Intern. Med. 2017, 31, 1771–1781. [Google Scholar] [CrossRef]
- Dewey, C.W.; Marino, D.J.; Loughin, C.A. Craniocervical junction abnormalities in dogs. N. Z. Vet. J. 2013, 61, 202–211. [Google Scholar] [CrossRef]
- Speer, M.C.; Enterline, D.S.; Mehltretter, L.; Hammock, P.; Joseph, J.; Dickerson, M.; Ellenbogen, R.G.; Milhorat, T.H.; Hauser, M.A.; George, T.M. Review Article: Chiari Type I Malformation with or Without Syringomyelia: Prevalence and Genetics. J. Genet. Couns. 2003, 12, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Bodensteiner, J.B. Neurological Manifestations of Achondroplasia. Curr. Neurol. Neurosci. Rep. 2019, 19, 105. [Google Scholar] [CrossRef] [PubMed]
- Giner, J.; Pérez López, C.; Hernández, B.; Gómez de la Riva, A.; Isla, A.; Roda, J.M. Update on the pathophysiology and management of syringomyelia unrelated to Chiari malformation. Neurologia 2019, 34, 318–325. [Google Scholar] [CrossRef]
- Rusbridge, C. New considerations about Chiari-like malformation, syringomyelia and their management. Practice 2020, 42, 252–267. [Google Scholar] [CrossRef]
- Lewis, T.; Rusbridge, C.; Knowler, P.; Blott, S.; Woolliams, J.A. Heritability of syringomyelia in Cavalier King Charles spaniels. Vet. J. 2010, 183, 345–347. [Google Scholar] [CrossRef]
- Lemay, P.; Knowler, S.P.; Bouasker, S.; Nédélec, Y.; Platt, S.; Freeman, C.; Child, G.; Barreiro, L.; Rouleau, G.A.; Rusbridge, C.; et al. Quantitative Trait Loci (QTL) Study Identifies Novel Genomic Regions Associated to Chiari-Like Malformation in Griffon Bruxellois Dogs. PLoS ONE 2014, 9, e89816. [Google Scholar] [CrossRef]
- Ancot, F.; Lemay, P.; Knowler, S.P.; Kennedy, K.; Griffiths, S.; Cherubini, G.B.; Sykes, J.; Mandigers, P.J.J.; Rouleau, G.A.; Rusbridge, C.; et al. A genome-wide association study identifies candidate loci associated to syringomyelia secondary to Chiari-like malformation in Cavalier King Charles Spaniels. BMC Genet. 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Mayousse, V.; Desquilbet, L.; Jeandel, A.; Blot, S. Prevalence of neurological disorders in French bulldog: A retrospective study of 343 cases (2002–2016). BMC Vet. Res. 2017, 13, 212. [Google Scholar] [CrossRef]
- Ryan, R.; Gutierrez-Quintana, R.; ter Haar, G.; De Decker, S. Prevalence of thoracic vertebral malformations in French bulldogs, Pugs and English bulldogs with and without associated neurological deficits. Vet. J. 2017, 221, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ricco, C.F.; Samarani, F.; Behr, S.; Gomes, E.; Cauzinille, L. Significance of suspected acquired cervical syringomyelia in 12 French Bulldogs with neurological deficits. Rev. Med. Vet. 2018, 169, 209–216. [Google Scholar]
- Mukherjee, S.; Simon, J.; Bayuga, S.; Ludwig, E.; Yoo, S.; Orlow, I.; Viale, A.; Offit, K.; Kurtz, R.C.; Olson, S.H.; et al. Including Additional Controls from Public Databases Improves the Power of a Genome-Wide Association Study. Hum. Hered. 2011, 72, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformáticas 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Garrison, E.; Kronenberg, Z.N.; Dawson, E.T.; Pedersen, B.S.; Prins, P. Vcflib and tools for processing the VCF variant call format. BioRxiv 2021. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef]
- Smith, P.J.; Zhang, C.; Wang, J.; Chew, S.L.; Zhang, M.Q.; Krainer, A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006, 15, 2490–2508. [Google Scholar] [CrossRef]
- Fondon, J.W.; Garner, H.R. Detection of length-dependent effects of tandem repeat alleles by 3-D geometric decomposition of craniofacial variation. Dev. Genes Evol. 2006, 217, 79–85. [Google Scholar] [CrossRef]
- Marchant, T.W.; Johnson, E.J.; McTeir, L.; Johnson, C.I.; Gow, A.; Liuti, T.; Kuehn, D.; Svenson, K.; Bermingham, M.L.; Drögemüller, M.; et al. Canine Brachycephaly Is Associated with a Retrotransposon-Mediated Missplicing of SMOC2. Curr. Biol. 2017, 27, 1573–1584. [Google Scholar] [CrossRef]
- Schoenebeck, J.J.; Hutchinson, S.A.; Byers, A.; Beale, H.C.; Carrington, B.; Faden, D.L.; Rimbault, M.; Decker, B.; Kidd, J.M.; Sood, R.; et al. Variation of BMP3 Contributes to Dog Breed Skull Diversity. PLoS Genet. 2012, 8, e1002849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, E.A.; Dickinson, P.J.; Mansour, T.; Sturges, B.K.; Aguilar, M.; Young, A.E.; Korff, C.; Lind, J.; Ettinger, C.L.; Varon, S.; et al. FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs. Proc. Natl. Acad. Sci. USA 2017, 114, 11476–11481. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, P.J.; Bannasch, D.L. Current Understanding of the Genetics of Intervertebral Disc Degeneration. Front. Veter. Sci. 2020, 7, 431. [Google Scholar] [CrossRef]
- Mansour, T.A.; Lucot, K.; Konopelski, S.E.; Dickinson, P.J.; Sturges, B.K.; Vernau, K.L.; Choi, S.; Stern, J.A.; Thomasy, S.M.; Döring, S.; et al. Whole genome variant association across 100 dogs identifies a frame shift mutation in DISHEVELLED 2 which contributes to Robinow-like syndrome in Bulldogs and related screw tail dog breeds. PLoS Genet. 2018, 14, e1007850. [Google Scholar] [CrossRef] [PubMed]
- Niskanen, J.E.; Reunanen, V.; Salonen, M.; Bannasch, D.; Lappalainen, A.K.; Lohi, H.; Hytönen, M.K. Canine DVL2 variant contributes to brachycephalic phenotype and caudal vertebral anomalies. Qual. Life Res. 2021, 140, 1535–1545. [Google Scholar] [CrossRef]
- Hayashi, M.; Nakashima, T.; Taniguchi, M.; Kodama, T.; Kumanogoh, A.; Takayanagi, H. Osteoprotection by semaphorin 3A. Nature 2012, 485, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Bernard, F.; Moreau-Fauvarque, C.; Heitz-Marchaland, C.; Zagar, Y.; Dumas, L.; Fouquet, S.; Lee, X.; Shao, Z.; Mi, S.; Chédotal, A. Role of transmembrane semaphorin Sema6A in oligodendrocyte differentiation and myelination. Glia 2012, 60, 1590–1604. [Google Scholar] [CrossRef]
- Shim, S.-O.; Cafferty, W.B.; Schmidt, E.C.; Kim, B.G.; Fujisawa, H.; Strittmatter, S.M. PlexinA2 limits recovery from corticospinal axotomy by mediating oligodendrocyte-derived Sema6A growth inhibition. Mol. Cell. Neurosci. 2012, 50, 193–200. [Google Scholar] [CrossRef]
- Oh, J.-E.; Kim, H.J.; Kim, W.-S.; Lee, Z.H.; Ryoo, H.-M.; Hwang, S.J.; Lee, Y.; Kim, H.-H. PlexinA2 mediates osteoblast differentiation via regulation of Runx2. J. Bone Miner. Res. 2011, 27, 552–562. [Google Scholar] [CrossRef]
- Kajii, T.S.; Oka, A.; Hatta, M.; Yamazaki, J.; Yamashita, J.; Iida, J. PLXNA2 identified as a candidate gene by genome-wide association analysis for mandibular prognathism in human chondrocytes. Biomed. Rep. 2018, 9, 253–258. [Google Scholar] [CrossRef]
- Lewandowska, M.A. The missing puzzle piece: Splicing mutations. Int. J. Clin. Exp. Pathol. 2013, 6, 2675–2682. [Google Scholar] [PubMed]
- Abdel-Salam, G.M.H.; Mazen, I.; Eid, M.; Ewida, N.; Shaheen, R.; Alkuraya, F.S. Biallelic novel missense HHAT variant causes syndromic microcephaly and cerebellar-vermis hypoplasia. Am. J. Med. Genet. Part A 2019, 179, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.F.; Kurosaka, H.; Iulianella, A.; Pace, J.; Thomas, N.; Beckham, S.; Williams, T.; Trainor, P.A. Mutations in Hedgehog Acyltransferase (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects. PLoS Genet. 2012, 8, e1002927. [Google Scholar] [CrossRef] [PubMed]
- Belloni, E.; Muenke, M.; Roessler, E.; Traverse, G.; Siegel-Bartelt, J.; Frumkin, A.; Mitchell, H.; Donis-Keller, H.; Helms, C.; Hing, A.; et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 1996, 14, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; De Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; De Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, A.M.; Racacho, L.; Grimsey, A.; Hudgins, L.; Kwan, A.C.; Sangalli, M.; Kidd, A.; Yaron, Y.; Lau, Y.-L.; Nikkel, S.M.; et al. Brachydactyly A-1 mutations restricted to the central region of the N-terminal active fragment of Indian Hedgehog. Eur. J. Hum. Genet. 2009, 17, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Scherz, P.J.; Harfe, B.D.; McMahon, A.P.; Tabin, C.J. The Limb Bud Shh-Fgf Feedback Loop Is Terminated by Expansion of Former ZPA Cells. Science 2004, 305, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Jean, P.H.; Hugnot, J.P.; Franzen, R. The spinal cord ependymal region: A stem cell niche in the caudal central nervous system. Front. Biosci. 2011, 16, 1044–1059. [Google Scholar] [CrossRef]
- Tabe, S.; Hikiji, H.; Ariyoshi, W.; Hashidate-Yoshida, T.; Shindou, H.; Shimizu, T.; Okinaga, T.; Seta, Y.; Tominaga, K.; Nishihara, T. Lysophosphatidylcholine acyltransferase 4 is involved in chondrogenic differentiation of ATDC5 cells. Sci. Rep. 2017, 7, 16701. [Google Scholar] [CrossRef]
- Wang, P.; Ma, K.; Yang, L.; Zhang, G.; Ye, M.; Wang, S.; Wei, S.; Chen, Z.; Gu, J.; Zhang, L.; et al. Predicting signaling pathways regulating demyelination in a rat model of lithium-pilocarpine-induced acute epilepsy: A proteomics study. Int. J. Biol. Macromol. 2021, 193, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Schmied, M.C.; Zehetmayer, S.; Reindl, M.; Ehling, R.; Bajer-Kornek, B.; Leutmezer, F.; Zebenholzer, K.; Hotzy, C.; Lichtner, P.; Meitinger, T.; et al. Replication study of multiple sclerosis (MS) susceptibility alleles and correlation of DNA-variants with disease features in a cohort of Austrian MS patients. Neurogenetics 2012, 13, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Bader, B.L.; Rayburn, H.; Crowley, D.; Hynes, R.O. Extensive Vasculogenesis, Angiogenesis, and Organogenesis Precede Lethality in Mice Lacking All αv Integrins. Cell 1998, 95, 507–519. [Google Scholar] [CrossRef]
- Mori, S.; Hatori, N.; Kawaguchi, N.; Hamada, Y.; Shih, T.-C.; Wu, C.-Y.; Lam, K.S.; Matsuura, N.; Yamamoto, H.; Takada, Y.K.; et al. The integrin-binding defective FGF2 mutants potently suppress FGF2 signalling and angiogenesis. Biosci. Rep. 2017, 37, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, J.; Hegde, A.; Andrade, A.C.; Nilsson, O.; Baron, J. Fibroblast growth factor expression in the postnatal growth plate. Bone 2007, 40, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Kyöstilä, K.; Lappalainen, A.K.; Lohi, H. Canine Chondrodysplasia Caused by a Truncating Mutation in Collagen-Binding Integrin Alpha Subunit 10. PLoS ONE 2013, 8, e75621. [Google Scholar] [CrossRef] [PubMed]
- Gianni, T.; Salvioli, S.; Chesnokova, L.S.; Hutt-Fletcher, L.M.; Campadelli-Fiume, G. αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion. PLoS Pathog. 2013, 9, e1003806. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, D.Y.; Huong, S.-M.; Huang, E.-S. Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 2005, 11, 515–521. [Google Scholar] [CrossRef]
- Bruhn, S.; Katzenellenbogen, M.; Gustafsson, M.; Krönke, A.; Sönnichsen, B.; Zhang, H.; Benson, M. Combining gene expression microarray- and cluster analysis with sequence-based predictions to identify regulators of IL-13 in allergy. Cytokine 2012, 60, 736–740. [Google Scholar] [CrossRef] [PubMed]
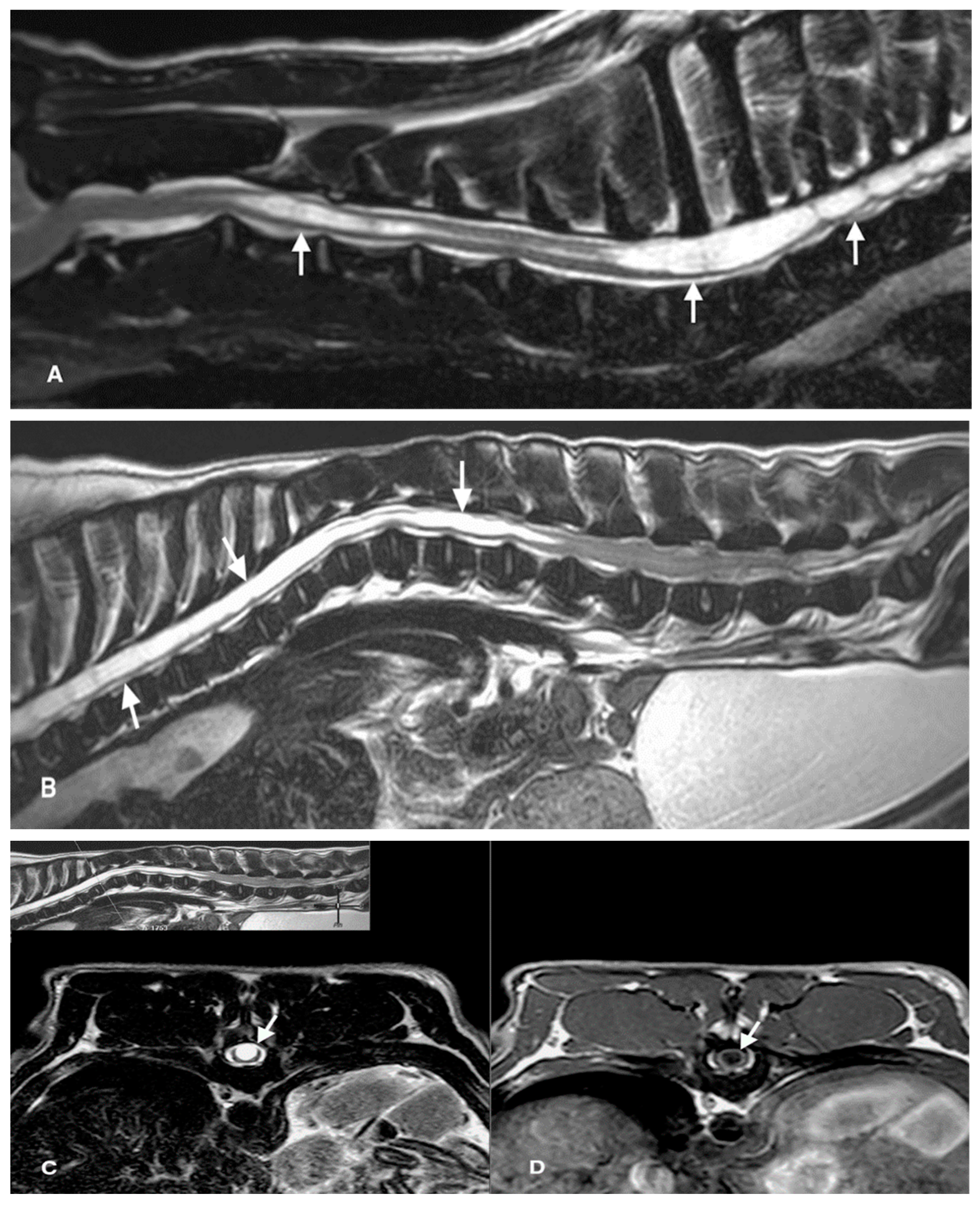
| Group | Dog | Sex | Age (Years) | Weight (kg) | Location of SHM | Neurological Dysfunction a | Intervertebral Disc Disease | Spinal Malformation b | Spondyloarthrosis | Another Disease |
|---|---|---|---|---|---|---|---|---|---|---|
| SHM cases | 1-40353 | F | 10.6 | 12.2 | Cervical, thoracic, lumbar | Grade II | C2-C3, T11-T12, T12-T13, T13-L1 | VM T3, T4, T7, T11 | No | Brachycephalic syndrome |
| 2-40354 | M | 3.2 | 12 | Cervical | Grade I | C3-C4 | VM T5-T7 | No | Brachycephalic syndrome | |
| 3-40462 | M | 4.6 | 15.8 | Cervical, thoracic, lumbar | Grade II | No | VM T5-T6 | L7-S1 | Stones and recurrent cystitis | |
| 4-40468 | F | 7.2 | 11.9 | Thoracic, lumbar | Asymptomatic | L6-L7, L5-L6, L7-S1 | VM T4-T9 | No | Heart murmur, allergies | |
| 5-38819 | F | 9.9 | 11.8 | Cervical | Grade II | C4-C5, T12-T13, L3-L4, L7-S1 | VM T3-T12 | Generalized | Pyometra, mammary nodules, allergies | |
| 6-38818 | F | 4.5 | 9 | Cervical, thoracic, lumbar | Grade II | C2-C3, C3-C4, C4-C5, T13-L1, L7-S1 | VM T2, T6, T8 | L7-S1 | No | |
| 7-38814 | M | 5.4 | 13 | Thoracic | Grade II | T11-T12, T13-L1, L1-L2, L2-L3, L4-L5, L7-S1 | VM T5-T9, meningocele L7 | No | Cystitis | |
| 8-39789 | M | 11.3 | 11 | Cervical, thoracic, lumbar | Grade III | C4-C5, C5-C6, T12-T13, T13-L1, L1-L2, L4-L5, L7-S1 | VM T2-T10 | L7-S1 | Scrotal tumor | |
| 9-38127 | F | 7.5 | - | Cervical | Grade II | C2-C3, C3-C4, C4-C5, C5-C6, C6-C7 | No | No | Allergies | |
| 10-38811 | M | 10.2 | 10.2 | Cervical | Grade III | C3-C4 | No | No | Brain neoplasia (no biopsy) | |
| 11-38212 | M | 4.9 | 10.9 | Cervical | Grade I | C6-C7 | No | No | No | |
| 12-38211 | F | 11.9 | 14.8 | Cervical | Grade II | No | No | Lumbar | Recurrent gastritis, neoplasia (C5 lesion with inconclusive biopsy) | |
| Common controls | 30-38213 | M | 5.4 | 16 | - | - | C4-C5, C5-C6, C6-C7, L7-S1 | No | No | No |
| 31-38215 | F | 9.4 | 11.5 | - | - | T5-T6, L1-L2, L2-L3 | VM T5, leptomeningeal cavitation T11 | No | Dislocation of the right patella, osteoarthritis in the hips, gums nodules | |
| 32-40466 | M | 7 | 9.7 | - | - | C2-C3, C5-C6, C6-C7 | VM T6-T10 | T9-T10, T12-T13, L7-S1 | No | |
| 33-40357 | F | 5 | 16.2 | - | - | L7-S1 | VM T4-T8 | No | No | |
| 34-40469 | M | 7.8 | 12 | - | - | C2-C3, C5-C6, C6-C7, L4-L5, L7-S1 | VM T5-T12 | T5-T7, L7-S1 | No | |
| 35-40471 | F | 12 | 9.7 | - | - | C4-C5, C6-C7, L1-L2, L3-L4, L4-L5, L7-S1 | No | L7-S1 | Biliary cholestasis, splenic neo/hyperplasia, renal cortical cysts, heart murmur | |
| 36-40464 | F | 6.9 | 11.2 | - | - | C2-C3, C3-C4, L1-L2, L2-L3 | VM T5-T10 | No | No | |
| 37-39790 | M | 6.6 | 12 | - | - | C2-C3, C3-C4, T12-T13, L4-L5, L7-S1 | VM T3-T13 | L7-S1 | No | |
| 38-38816 | F | 5 | 9.8 | - | - | C2-C3, C4-C5, C5-C6, L1-L2, L7-S1 | VM T3, T5, T6, T9, T13, L6, meningocele L5-L7 | No | No | |
| 39-38217 | M | 5 | 15 | - | - | C3-C4, L6-L7, L7-S1 | No | No | No |
| Position/Region in Canfam 3.1 Assembly | Gene Name and Symbol | Candidate Variants | |||
|---|---|---|---|---|---|
| Alleles (ref/alt) | Position | p Value | Gene Location/Effect | ||
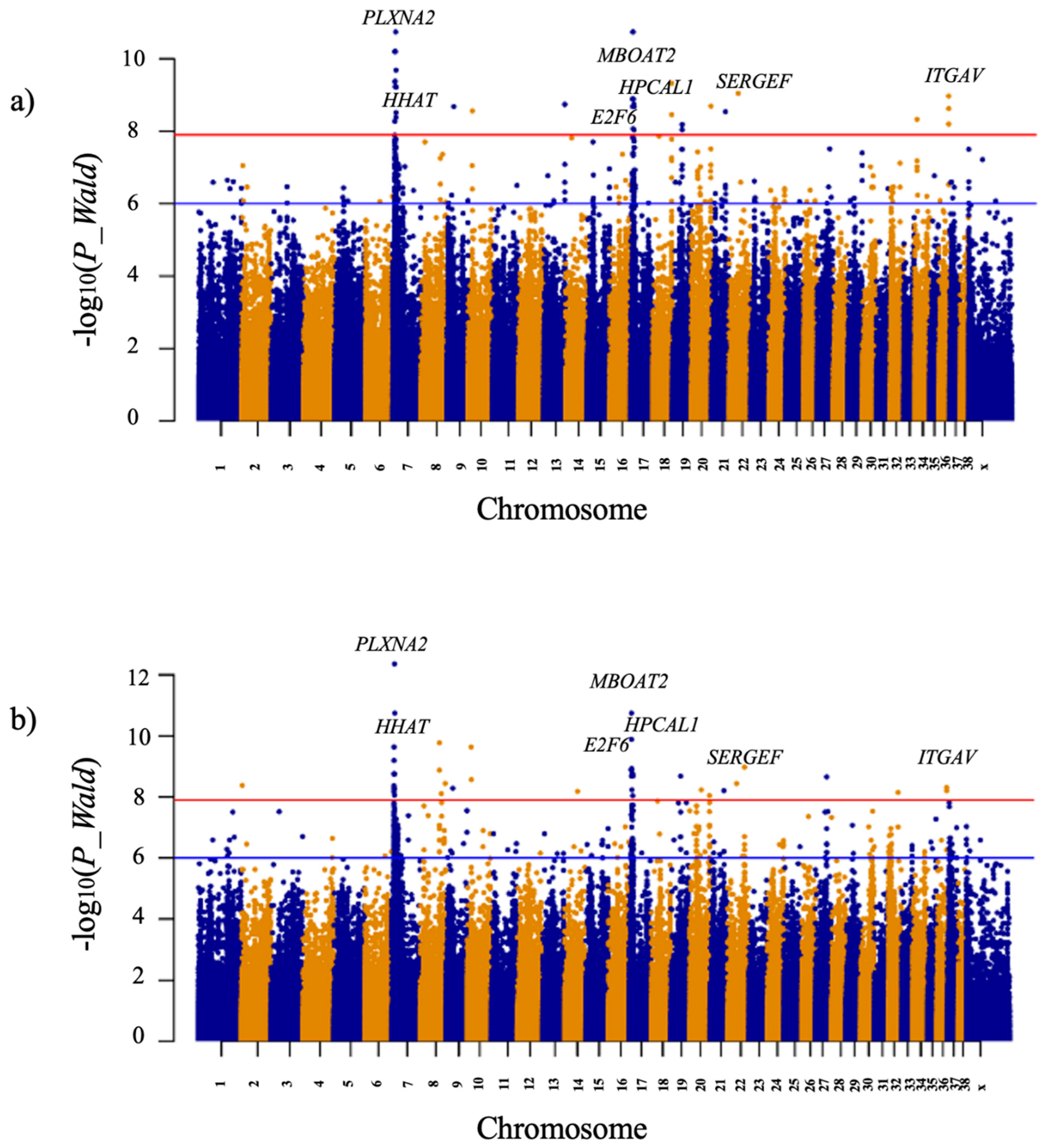
| CFA7:6938927-7043294 | plexin A2 (PLXNA2) | T/C | 7043294 | 6.23 × 10−11 | Intronic/Branch Site mutation |
| CFA7:9012453 | hedgehog acyltransferase (HHAT) | InsT | 9012453 | 1.80 × 10−11 | Intronic |
| CFA17:6251280 | membrane bound O-acyltransferase domain containing 2 (MBOAT2) | G/A | 6251280 | 1.28 × 10−9 | Upstream 5′UTR |
| CFA17:7242110 | hippocalcin like 1 (HPCAL1) | C/T | 7242110 | 1.80 × 10−11 | Upstream 5′UTR |
| CFA17:8153564 | E2F transcription factor 6 (E2F6) | A/G | 8153564 | 1.28 × 10−9 | Downstream 3′UTR |
| CFA21:40329686 | secretion regulating guanine nucleotide exchange factor (SERGEF) | C/A | 40329686 | 2.88 × 10−9 | Intronic/Branch Site mutation |
| CFA36:28842415-28868657 | integrin subunit alpha V (ITGAV) | A/G | 28842415 | 6.29 × 10−9 | Intronic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrino, S.; Lorenzo, V.; Dunner, S.; Contreras, E.; Cañón, J.; Sevane, N. Syringohydromyelia in Dogs: The Genomic Component Underlying a Complex Neurological Disease. Animals 2022, 12, 2622. https://doi.org/10.3390/ani12192622
Andrino S, Lorenzo V, Dunner S, Contreras E, Cañón J, Sevane N. Syringohydromyelia in Dogs: The Genomic Component Underlying a Complex Neurological Disease. Animals. 2022; 12(19):2622. https://doi.org/10.3390/ani12192622
Chicago/Turabian StyleAndrino, Sandra, Valentina Lorenzo, Susana Dunner, Elisabeth Contreras, Javier Cañón, and Natalia Sevane. 2022. "Syringohydromyelia in Dogs: The Genomic Component Underlying a Complex Neurological Disease" Animals 12, no. 19: 2622. https://doi.org/10.3390/ani12192622






