Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
2.3. Bacterial Isolation
2.4. Phenotypic Determination of Antimicrobial Resistance
2.5. Whole Genome Sequencing and Identification of Antimicrobial Resistance Genes
2.6. Statistical Analysis
3. Results
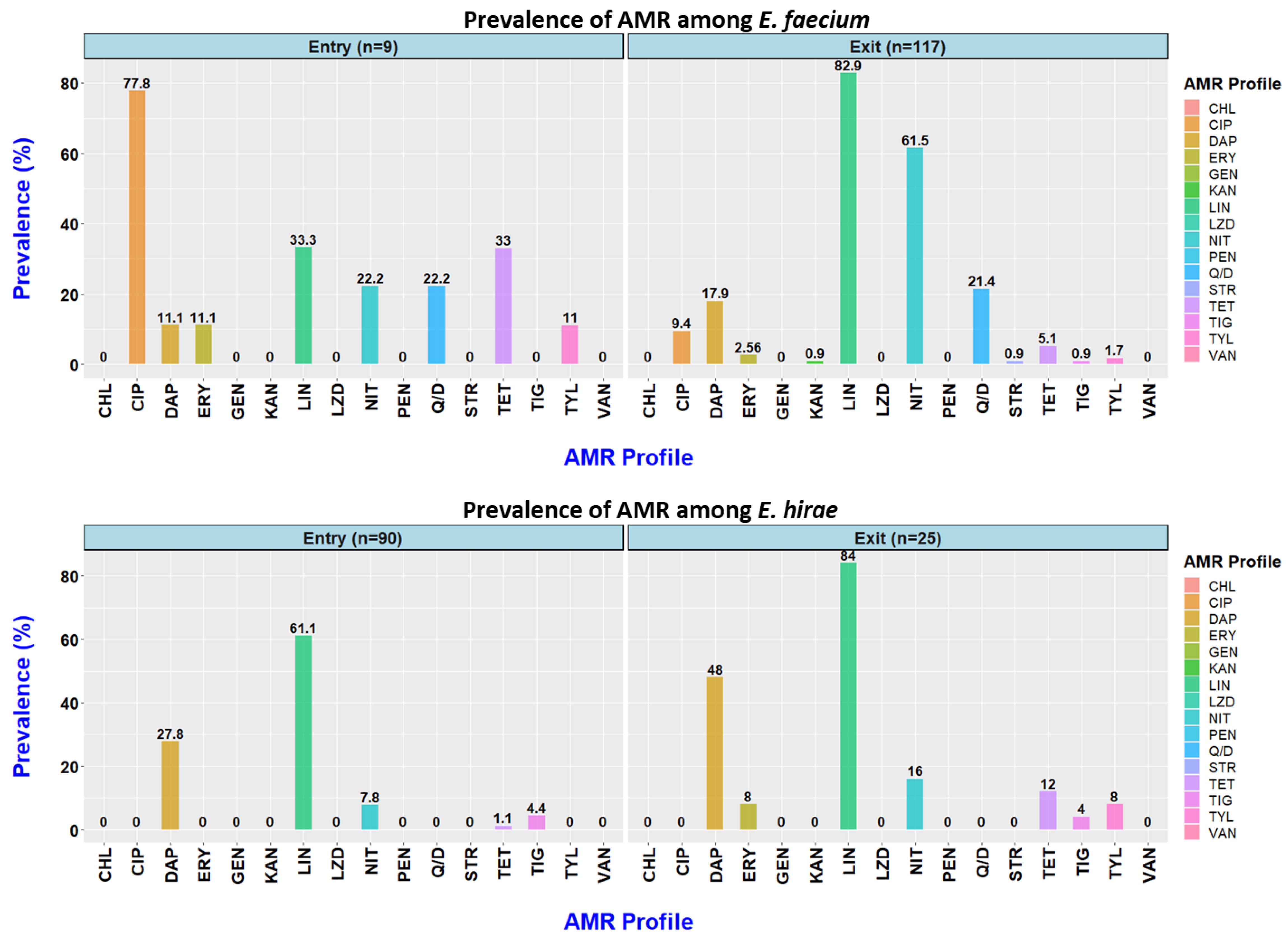
3.1. Prevalence of Antimicrobial Resistance at Entry into the Feedlot
3.2. Prevalence of AMR at Exit from the Feedlot
3.3. Antimicrobial Resistance Profiles
3.4. Changes in Antimicrobial Resistance Status Observed among Enterococcus faecium and E. hirae between Feedlot Entry and Exit
3.5. Antimicrobial Resistance Genes Identified among Enterococcus faecium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirchhelle, C. Pharming animals: A global history of antibiotics in food production (1935–2017). Palgrave Commun. 2018, 4, 96. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Scott, A.M.; Beller, E.; Glasziou, P.; Clark, J.; Ranakusuma, R.W.; Byambasuren, O.; Bakhit, M.; Page, S.W.; Trott, D.; Mar, C.D. Is antimicrobial administration to food animals a direct threat to human health? A rapid systematic review. Int. J. Antimicrob. Agents 2018, 52, 316–323. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.O.; Dantas, G. Antibiotics and the resistant microbiome. Curr. Opin. Microbiol. 2011, 14, 556–563. [Google Scholar] [CrossRef]
- Boehm, A.B.; Sassoubre, L.M. Enterococci as indicators of environmental fecal contamination. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Sherman, J.M. The Enterococci and Related Streptococci. J. Bacteriol. 1938, 35, 81–93. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef]
- Barlow, R.S.; McMillan, K.E.; Duffy, L.L.; Fegan, N.; Jordan, D.; Mellor, G.E. Antimicrobial resistance status of Enterococcus from Australian cattle populations at slaughter. PLoS ONE 2017, 12, e0177728. [Google Scholar] [CrossRef]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef]
- Kos, V.N.; Desjardins, C.A.; Griggs, A.; Cerqueira, G.; Van Tonder, A.; Holden, M.T.; Godfrey, P.; Palmer, K.L.; Bodi, K.; Mongodin, E.F.; et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 2012, 3, e00112-12. [Google Scholar] [CrossRef]
- de Jong, A.; Simjee, S.; Garch, F.E.; Moyaert, H.; Rose, M.; Youala, M.; Dry, M. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics. Vet. Microbiol. 2018, 216, 168–175. [Google Scholar] [CrossRef]
- Hermanovská, L.; Bardoň, J.; Čermák, P. Vancomycin-resistant enterococci—The nature of resistance and risk of transmission from animals to humans. Klin. Mikrobiol. Infekc. Lek. 2016, 22, 54–60. [Google Scholar] [PubMed]
- Olsen, R.H.; Schønheyder, H.C.; Christensen, H.; Bisgaard, M. Enterococcus faecalis of Human and Poultry Origin Share Virulence Genes Supporting the Zoonotic Potential of E. faecalis. Zoonoses Public Health 2012, 59, 256–263. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef] [PubMed]
- O’Dea, M.; Sahibzada, S.; Jordan, D.; Laird, T.; Lee, T.; Hewson, K.; Pang, S.; Abraham, R.; Coombs, G.W.; Harris, T.; et al. Genomic, Antimicrobial Resistance, and Public Health Insights into Enterococcus spp. from Australian Chickens. J. Clin. Microbiol. 2019, 57, e00319-19. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.H.; Hwang, S.Y.; Moon, B.Y.; Park, Y.K.; Shin, S.; Hwang, C.-Y.; Park, Y.H. Occurrence of antimicrobial resistance and virulence genes, and distribution of enterococcal clonal complex 17 from animals and human beings in Korea. J. Vet. Diagn. Investig. 2012, 24, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, K.; Lai, J.; Wu, C.; Shen, J.; Wang, Y. Prevalence and antimicrobial resistance of Enterococcus species of food animal origin from Beijing and Shandong Province, China. J. Appl. Microbiol. 2013, 114, 555–563. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Jordan, D.; Sahibzada, S.; Abraham, R.; Pang, S.; Coombs, G.W.; O’Dea, M.; Abraham, S. Antimicrobial Resistance in Porcine Enterococci in Australia and the Ramifications for Human Health. Appl. Environ. Microbiol. 2021, 87, e03037-20. [Google Scholar] [CrossRef]
- Vignaroli, C.; Zandri, G.; Aquilanti, L.; Pasquaroli, S.; Biavasco, F. Multidrug-resistant enterococci in animal meat and faeces and co-transfer of resistance from an Enterococcus durans to a human Enterococcus faecium. Curr. Microbiol. 2011, 62, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- FDA. National Antimicrobial Resistance Monitoring System. Retail Meat Annual Report; FDA: Silver Spring, MD, USA, 2011. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. 2010. 2017. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 January 2022).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017, 15, e04694. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar] [CrossRef]
- van Hal, S.J.; Ip, C.L.C.; Ansari, M.A.; Wilson, D.J.; Espedido, B.A.; Jensen, S.O.; Bowden, R. Evolutionary dynamics of Enterococcus faecium reveals complex genomic relationships between isolates with independent emergence of vancomycin resistance. Microb. Genom. 2016, 2, e000048. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal gut bacteria: Distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 2012, 62, 449–459. [Google Scholar] [CrossRef]
- Ramos, S.; Igrejas, G.; Capelo-Martinez, J.-L.; Poeta, P. Antibiotic resistance and mechanisms implicated in fecal enterococci recovered from pigs, cattle and sheep in a Portuguese slaughterhouse. Ann. Microbiol. 2012, 62, 1485–1494. [Google Scholar] [CrossRef]
- ASTAG. Importance Ratings and Summary of Antibacterial Uses in Human and Animal Health in Australia; Version 1.0; Australian Strategic and Technical Advisory Group on AMR (ASTAG): Canberra, Australia, 2018. [Google Scholar]
- Argudín, M.A.; Deplano, A.; Meghraoui, A.; Dodémont, M.; Heinrichs, A.; Denis, O.; Nonhoff, C.; Roisin, S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics 2017, 6, 12. [Google Scholar] [CrossRef]
- Devriese, L.A.; Laurier, L.; De Herdt, P.; Haesebrouck, F. Enterococcal and streptococcal species isolated from faeces of calves, young cattle and dairy cows. J. Appl. Bacteriol. 1992, 72, 29–31. [Google Scholar] [CrossRef]
- Johnston, L.M.; Jaykus, L.-A. Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 2004, 70, 3133–3137. [Google Scholar] [CrossRef]
- Tyson, G.; Nyirabahizi, E.; Crarey, E.; Kabera, C.; Lam, C.; Rice-Trujillo, C.; McDermott, P.; Tate, H. Prevalence and Antimicrobial Resistance of Enterococci Isolated from Retail Meats in the United States, 2002–2014. Appl. Environ. Microbiol. 2017, 84, e01902-17. [Google Scholar] [CrossRef] [PubMed]
- Barlow, R.; McMillan, K.; Abraham, S. Antimicrobial Resistance in Commensal Bacteria in Bovine Faeces at Slaughter; Meat and Livestock Australia Limited: North Sydney, Australia, 2020. [Google Scholar]
- Perry, F.G. Biotechnology in animal feeds and animal feeding: An overview. In Biotechnology in Animal Feeds and Animal Feeding; VCH Verlagsgesellschaft mbH: Weinhaim, Germany, 1995; pp. 1–15. [Google Scholar]
- Mikalsen, T.; Pedersen, T.; Willems, R.; Coque, T.M.; Werner, G.; Sadowy, E.; van Schaik, W.; Jensen, L.B.; Sundsfjord, A.; Hegstad, K. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 2015, 16, 282. [Google Scholar] [CrossRef] [PubMed]
- Beukers, A.G.; Zaheer, R.; Goji, N.; Amoako, K.K.; Chaves, A.V.; Ward, M.P.; McAllister, T.A. Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. 2017, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Faron, M.L.; Ledeboer, N.A.; Buchan, B.W. Resistance Mechanisms, Epidemiology, and Approaches to Screening for Vancomycin-Resistant Enterococcus in the Health Care Setting. J. Clin. Microbiol. 2016, 54, 2436. [Google Scholar] [CrossRef] [PubMed]
- Arsène, S.; Leclercq, R. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 2007, 51, 3254–3258. [Google Scholar] [CrossRef]
- Jonas, B.M.; Murray, B.E.; Weinstock, G.M. Characterization of emeA, a NorA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob. Agents Chemother. 2001, 45, 3574–3579. [Google Scholar] [CrossRef]
- Kim, Y.B.; Seo, H.J.; Seo, K.W.; Jeon, H.Y.; Kim, D.K.; Kim, S.W.; Lim, S.-K.; Lee, Y.J. Characteristics of High-Level Ciprofloxacin-Resistant Enterococcus faecalis and Enterococcus faecium from Retail Chicken Meat in Korea. J. Food Prot. 2018, 81, 1357–1363. [Google Scholar] [CrossRef]
- Wetzstein, H.G. Comparative mutant prevention concentrations of pradofloxacin and other veterinary fluoroquinolones indicate differing potentials in preventing selection of resistance. Antimicrob. Agents Chemother. 2005, 49, 4166–4173. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updates 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Arias, C.A.; Panesso, D.; McGrath, D.M.; Qin, X.; Mojica, M.F.; Miller, C.; Diaz, L.; Tran, T.T.; Rincon, S.; Barbu, E.M.; et al. Genetic Basis for In Vivo Daptomycin Resistance in Enterococci. N. Engl. J. Med. 2011, 365, 892–900. [Google Scholar] [CrossRef]
- Jancel, T.; Dudas, V. Management of uncomplicated urinary tract infections. West J. Med. 2002, 176, 51–55. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Peixe, L. Illegal use of nitrofurans in food animals: Contribution to human salmonellosis? Clin. Microbiol. Infect. 2006, 12, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Shakti, L.; Veeraraghavan, B. Advantage and limitations of nitrofurantoin in multi-drug resistant Indian scenario. Indian J. Med. Microbiol. 2015, 33, 477–481. [Google Scholar] [CrossRef]
- Ho, P.L.; Ng, K.Y.; Lo, W.U.; Law, P.Y.; Lai, E.L.; Wang, Y.; Chow, K.H. Plasmid-Mediated OqxAB Is an Important Mechanism for Nitrofurantoin Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 537–543. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Zhou, C.; Lin, Y.; Liu, S.; Zeng, W.; Yu, K.; Zhou, T.; Cao, J. Unraveling Mechanisms and Epidemic Characteristics of Nitrofurantoin Resistance in Uropathogenic Enterococcus faecium Clinical Isolates. Infect. Drug Resist. 2021, 14, 1601–1611. [Google Scholar] [CrossRef]
- Benedict, K.M.; Gow, S.P.; McAllister, T.A.; Booker, C.W.; Hannon, S.J.; Checkley, S.L.; Noyes, N.R.; Morley, P.S. Antimicrobial Resistance in Escherichia coli Recovered from Feedlot Cattle and Associations with Antimicrobial Use. PLoS ONE 2015, 10, e0143995. [Google Scholar] [CrossRef]

| Antimicrobial Agent | Range | Breakpoints for Resistance |
|---|---|---|
| Chloramphenicol | 2–32 | ≥32 a |
| Ciprofloxacin | 0.12–4 | ≥4 a |
| Daptomycin | 0.25–16 | ≥8 a |
| Erythromycin | 0.25–8 | ≥8 a |
| Gentamicin | 128–1024 | ≥512 b |
| Kanamycin | 128–1024 | ≥1024 b |
| Lincomycin | 1–8 | ≥8 b |
| Linezolid | 0.5–8 | ≥8 a |
| Nitrofurantoin | 2–64 | >64 a |
| Penicillin | 0.25–16 | ≥16 a |
| Streptomycin | 512–2048 | ≥1024 b |
| Quinupristin/Dalfopristin | 0.5–32 | ≥4 a |
| Tetracycline | 1–32 | ≥16 a |
| Tigecycline | 0.015–0.5 c | ≥0.5 b |
| Tylosine tartarte | 0.25–32 | ≥32 b |
| Vancomycin | 0.25–32 | ≥32 a |
| Antimicrobial Class | Antimicrobial Agent | Resistance (95% CI) | Percentage of Isolates Yielding Each MIC Value (µg/mL) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | |||
| Aminoglycosides | Gentamycin | 0.0 (0.00–3.56) | 100.0 | ||||||||||||||||
| Kanamycin | 0.0 (0.00–3.56) | 98.1 | 1.0 | 1.0 | |||||||||||||||
| Streptomycin | 0.0 (0.00–3.56) | 100.0 | |||||||||||||||||
| Beta-lactam | Penicillin | 0.0 (0.00–3.56) | 23.1 | 20.2 | 31.7 | 21.2 | 3.9 | ||||||||||||
| Fluoroquinolones | Ciprofloxacin | 6.7 (3.24–13.45) | 2.9 | 76.9 | 10.6 | 2.9 | 6.7 | ||||||||||||
| Glycopeptides | Vancomycin | 0.0 (0.00–3.56) | 1.0 | 50 | 48.1 | 1.0 | |||||||||||||
| Glycylcyclines | Tigecycline | 4.8 (2.02–11.03) | 14.4 | 43.3 | 29.8 | 7.7 | 4.8 | ||||||||||||
| Lincosamide | Lincomycin | 60.6 (50.91–69.48) | 36.5 | 1.0 | 1.9 | 5.8 | 54.8 | ||||||||||||
| Lipopeptides | Daptomycin | 25.0 (17.62–34.19) | 1.0 | 4.8 | 19.2 | 50.0 | 23.1 | 1.9 | |||||||||||
| Macrolides | Erythromycin | 1.0 (0.14–6.51) | 90.4 | 2.9 | 1.0 | 3.9 | 1.0 | 1.0 | |||||||||||
| Tylosine tartrate | 1.0 (0.14–6.51) | 1.0 | 3.9 | 24.0 | 57.7 | 11.5 | 1.0 | 1.0 | |||||||||||
| Nitrofurantoins | Nitrofurantoin | 8.65 (4.56–15.80) | 2.9 | 40.4 | 48.1 | 8.7 | |||||||||||||
| Oxazolidinones | Linezolid | 0.0 (0.00–3.56) | 1.0 | 3.9 | 94.2 | 1.0 | |||||||||||||
| Phenicols | Chloramphenicol | 0.0 (0.00–3.56) | 1.0 | 92.3 | 6.7 | ||||||||||||||
| Streptogramins | Quinupristin/dalfopristin | 2.9 (0.93–8.56) | 38.5 | 17.3 | 41.4 | 1.9 | 1.0 | ||||||||||||
| Tetracycline | Tetracycline | 3.9 (1.45–9.80) | 96.2 | 1.0 | 2.9 | ||||||||||||||
| Antimicrobial Class | Antimicrobial Agent | Resistance (95% CI) | Percentage of Isolates Yielding Each MIC Value (µg/mL) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | |||
| Aminoglycosides | Gentamycin | 0.0 (0.00–2.60) | 100.0 | ||||||||||||||||
| Kanamycin | 0.7 (0.10–4.76) | 70.1 | 25.7 | 3.5 | 0.7 | ||||||||||||||
| Streptomycin | 0.7 (0.10–4.76) | 99.3 | 0.7 | ||||||||||||||||
| Beta-lactam | Penicillin | 0.0 (0.00–2.60) | 8.3 | 12.5 | 11.1 | 23.6 | 43.8 | 0.7 | |||||||||||
| Fluoroquinolones | Ciprofloxacin | 7.6 (4.28–13.27) | 1.4 | 13.2 | 38.2 | 39.6 | 7.6 | ||||||||||||
| Glycopeptides | Vancomycin | 0.0 (0.00–2.60) | 60.4 | 34.0 | 4.2 | 1.4 | |||||||||||||
| Glycylcyclines | Tigecycline | 1.4 (0.35–5.38) | 0.7 | 3.5 | 52.1 | 38.2 | 4.2 | 1.4 | |||||||||||
| Lincosamide | Lincomycin | 84.0 (77.11–89.15) | 13.2 | 2.8 | 1.4 | 82.6 | |||||||||||||
| Lipopeptides | Daptomycin | 22.9 (16.77–30.48) | 1.4 | 12.5 | 63.2 | 22.2 | 0.7 | ||||||||||||
| Macrolides | Erythromycin | 4.2 (1.88–8.96) | 60.4 | 1.4 | 3.5 | 22.2 | 8.3 | 1.4 | 2.8 | ||||||||||
| Tylosine tartrate | 3.5 (1.45–8.07) | 0.7 | 25.7 | 24.3 | 45.8 | 3.5 | |||||||||||||
| Nitrofurantoins | Nitrofurantoin | 53.5 (45.30–61.46) | 3.5 | 43.1 | 53.5 | ||||||||||||||
| Oxazolidinones | Linezolid | 0.0 (0.00–2.60) | 0.7 | 97.2 | 2.1 | ||||||||||||||
| Phenicols | Chloramphenicol | 0.0 (0.00–2.60) | 10.4 | 89.6 | |||||||||||||||
| Streptogramins | Quinupristin/dalfopristin | 18.1 (12.59–25.20) | 13.2 | 4.2 | 64.6 | 17.4 | 0.7 | ||||||||||||
| Tetracycline | Tetracycline | 6.9 (3.78–12.43) | 93.1 | 0.7 | 6.3 | ||||||||||||||
| Antimicrobial Classes | Total No. of Isolates (%) | Resistance Pattern (No. of Isolates) | ||
|---|---|---|---|---|
| Entry (104) | Exit (144) | Entry | Exit | |
| All susceptible | 26 (25.00) | 4 (2.78) | 26 | 4 |
| 1 | 50 (48.08) | 44 (30.56) | LIN (38) | LIN (38) |
| DAP (6) | NIT (6) | |||
| CIP (3) | ||||
| TGC (2) | ||||
| TET (1) | ||||
| 2 | 18 (17.31) | 55 (38.19) | DAP-LIN (11) | LIN-NIT (28) |
| CIP-NIT (1) | LIN-Q/D (9) | |||
| CIP-TET (1) | CIP-LIN (1) | |||
| DAP-NIT (1) | DAP-NIT (7) | |||
| LIN-TIG (3) | DAP-LIN (7) | |||
| LIN-Q/D (1) | CIP-NIT (1) | |||
| LIN-TET (1) | ||||
| ERY-LIN-TYL (1) | ||||
| 3 | 8 (7.69) | 31 (21.53) | DAP-LIN-NIT (6) | LIN-NIT-Q/D (10) |
| CIP-LIN-NIT (1) | CIP-LIN-NIT (3) | |||
| DAP-LIN-TET (1) | CIP-DAP-NIT (3) | |||
| CIP-NIT-TIG (1) | ||||
| DAP-LIN-TET (2) | ||||
| DAP-LIN-Q/D (1) | ||||
| DAP-LIN-NIT (8) | ||||
| NIT-STR-TET (1) | ||||
| LIN-NIT-TET (1) | ||||
| ERY-LIN-TIG-TYL (1) | ||||
| 4 | 2 (1.92) | 9 (6.25) | CIP-DAP-LIN-Q/D (1) | DAP-LIN-NIT-TET (1) |
| ERY-LIN-Q/D-TET-TYL (1) | KAN-LIN-NIT-Q/D (2) | |||
| DAP-ERY-LIN-NIT (1) | ||||
| CIP-DAP-LIN-NIT (1) | ||||
| LIN-NIT-Q/D-TET (1) | ||||
| ERY-LIN-Q/D-TET-TYL (2) | ||||
| ERY-LIN-NIT-TET-TYL (1) | ||||
| 5 | 1 (0.69) | CIP-DAP-LIN-NIT-Q/D (1) | ||
| Non-MDR | 68 (65.4) | 99 (68.8) | ||
| MDR | 11 (10.6) | 41 (28.5) | ||
| Resistance | 79 (76.0) | 140 (96.3) | ||
| Antimicrobial Class | Resistance Phenotype | Resistance Gene | Number of Isolates (n = 62) |
|---|---|---|---|
| Aminoglycosides | GEN | aac(6′)-Ii | 59 (95.2) |
| Aminoglycosides | AMK | aac(6′)-Iid | 2 (3.2) |
| Aminoglycosides | STR | ant(6)-Ia | 1 (1.6) |
| β-lactam | AMP | pbp5 | 29 (46.8) |
| LsaP (lincosamides, streptogramin As and pleuromutilins) | Q/D, LIN | eatAv | 47 (75.8) |
| Lincosamide | LIN | lnu(G) | 2 (3.2) |
| Streptogramin | VIR, Q/D | vat -E | 1 (1.6) |
| Macrolide, streptogramin | ERY, Q/D, VIR | msr(C) | 59 (95.2) |
| MLS (macrolide, lincosamide, streptogramin) | ERY, LIN, Q/D | erm(B) | 3 (4.8) |
| Macrolides, fluoroquinolones | CIP | efmA | 21 (33.9) |
| Tetracyclines | TET | tet(M) | 3 (4.8) |
| Tetracyclines | TET | tet(L) | 2(3.2) |
| Tetracyclines | TET | tet(45) | 2(3.2) |
| Tetracyclines | TET | tet(S) | 2(3.2) |
| Antimicrobial Class | AMR Isolates (%) | Resistance Gene Observed (%) | Agreement (%) |
|---|---|---|---|
| Aminoglycosides | Kanamycin (n = 1; 1.6) | aac(6′)-Ii (n = 1;1.6) | 100 |
| Streptomycin (n = 0) | ant(6)-Ia (n = 1; 1.6) | 0 | |
| Fluoroquinolones | Ciprofloxacin (n = 18; 29.0) | efmA (n = 12; 19.3) | 66.7 |
| Lipopeptides | Daptomycin (n = 22; 35.5) | 0 | |
| Lincosamide | Lincomycin (n = 39; 62.9) | eatAv (n = 38; 61.3) | 97.4 |
| erm(B) (n = 3; 4.8) | |||
| lnu(G) (n = 2; 3.2) | |||
| Macrolides | Erythromycin (n = 4; 6.4) | msr(C) (n = 4; 6.4) | 100 |
| erm(B) (n = 3; 4.8) | |||
| Tylosin tartrate (n = 3; 4.8) | erm(B) (n = 3; 4.8) | 100 | |
| Nitrofurantoin | Nitrofurantoin (n = 27; 43.5) | 0 | |
| Streptogramins | Quinupristin/dalfopristin (n = 27; 43.5) | eatAv (n = 26; 41.9) | 96.3 |
| msr(C) (n = 26; 41.9) | |||
| Vat(E) (n = 1; 1.6) | |||
| Tetracycline | Tetracycline (n = 5; 8.1) | tet(M) (n = 3; 4.8) | 100 |
| tet(L) (n = 2; 3.2) | |||
| tet(S) (n = 2; 3.2) | |||
| tet(45) (n = 2; 3.2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messele, Y.E.; Hasoon, M.F.; Trott, D.J.; Veltman, T.; McMeniman, J.P.; Kidd, S.P.; Low, W.Y.; Petrovski, K.R. Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals 2022, 12, 2690. https://doi.org/10.3390/ani12192690
Messele YE, Hasoon MF, Trott DJ, Veltman T, McMeniman JP, Kidd SP, Low WY, Petrovski KR. Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals. 2022; 12(19):2690. https://doi.org/10.3390/ani12192690
Chicago/Turabian StyleMessele, Yohannes E., Mauida F. Hasoon, Darren J. Trott, Tania Veltman, Joe P. McMeniman, Stephen P. Kidd, Wai Y. Low, and Kiro R. Petrovski. 2022. "Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit" Animals 12, no. 19: 2690. https://doi.org/10.3390/ani12192690
APA StyleMessele, Y. E., Hasoon, M. F., Trott, D. J., Veltman, T., McMeniman, J. P., Kidd, S. P., Low, W. Y., & Petrovski, K. R. (2022). Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals, 12(19), 2690. https://doi.org/10.3390/ani12192690






