Sustainable Intensification of Beef Production in the Tropics: The Role of Genetically Improving Sexual Precocity of Heifers
Abstract
:Simple Summary
Abstract
1. Introduction
2. The Genetics of Heifers Sexual Precocity
2.1. Which Traits to Select?
2.1.1. Puberty Traits
2.1.2. Indicator Traits of Puberty
Scrotal Circumference (SC)
Age at First Conception (AFCo) or Calving (AFC)
Heifer Early Pregnancy (HP)
- Scenario 1
- Scenario 2
2.1.3. Sexual Precocity Evaluation Using High-Throughput Technologies
2.2. Genomic Selection
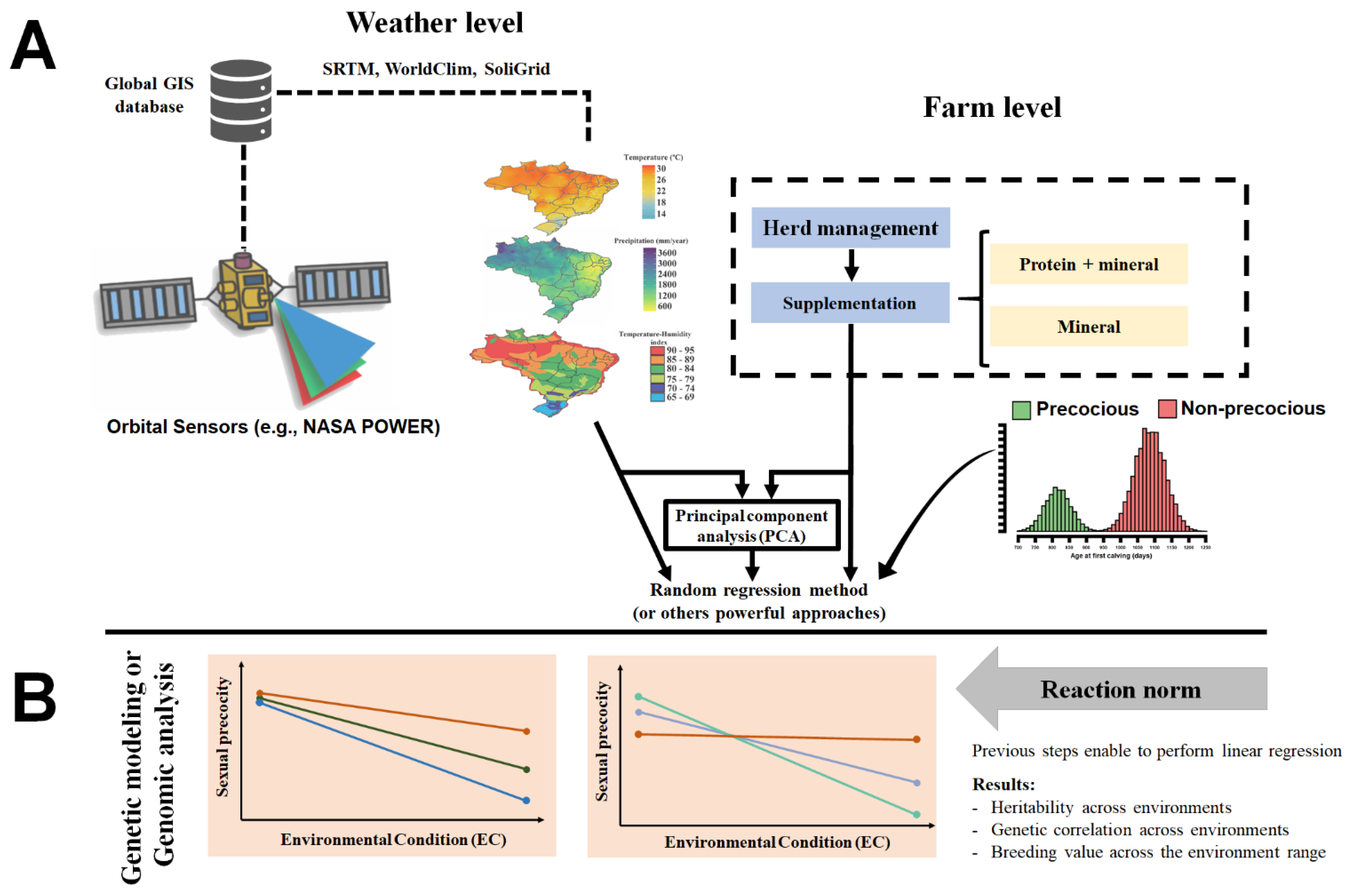
2.3. Genotype by Environment Interaction (G × E) for Sexual Precocity in Breeding Programs
G × E Interaction for Sexual Precocity in Genomic Era
2.4. Molecular Genetics
2.5. Case Study: Selection for Sexual Precocity in Nellore Breeding Programs
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albuquerque, L.G.; Fernandes Júnior, G.A.; Carvalheiro, R. Beef cattle genomic selection in tropical environments. In Proceedings of the 22nd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Townsville, Australia, 2–5 July 2017; Volume 22, pp. 255–263. [Google Scholar]
- Rudel, T.K.; Paul, B.; White, D.; Rao, I.M.; Van Der Hoek, R.; Castro, A.; Boval, M.; Lerner, A.; Schneider, L.; Peters, M. LivestockPlus: Forages, sustainable intensification, and food security in the tropics. Ambio 2015, 44, 685–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, B.; Snapp, S. What is sustainable intensification? Views from experts. Land Use Policy 2015, 46, 1–10. [Google Scholar] [CrossRef]
- Dumont, B.; Groot, J.C.J.; Tichit, M. Review: Make ruminants green again-how can sustainable intensification and agroecology converge for a better future? Animal 2018, 12, s210–s219. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.J.C.; Sorensen, J.T. Sustainability in cattle production systems. J. Agric. Environ. Ethics 1993, 6, 61–73. [Google Scholar] [CrossRef]
- Tedeschi, L.O.; Muir, J.P.; Riley, D.G.; Fox, D.G. The role of ruminant animals in sustainable livestock intensification programs. Int. J. Sustain. Dev. World Ecol. 2015, 22, 452–465. [Google Scholar] [CrossRef]
- Johnston, D.J.; Barwick, S.A.; Fordyce, G.; Holroyd, R.G.; Williams, P.J.; Corbet, N.J.; Grant, T. Genetics of early and lifetime annual reproductive performance in cows of two tropical beef genotypes in northern Australia. Anim. Prod. Sci. 2014, 54, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Johnston, D.J.; Bolormaa, S.; Hawken, R.J.; Tier, B. Genomic selection for female reproduction in Australian tropically adapted beef cattle. Anim. Prod. Sci. 2014, 54, 16–24. [Google Scholar] [CrossRef]
- Johnston, D.J.; Corbet, N.J.; Barwick, S.A.; Wolcott, M.L.; Holroyd, R.G. Genetic correlations of young bull reproductive traits and heifer puberty traits with female reproductive performance in two tropical beef genotypes in northern Australia. Anim. Prod. Sci. 2014, 54, 74–84. [Google Scholar] [CrossRef]
- Terakado, A.P.N.; Pereira, M.C.; Yokoo, M.J.; Albuquerque, L.G. Evaluation of productivity of sexually precocious Nelore heifers. Animal 2014, 9, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Júnior, G.A.; Garcia, D.A.; Hortolani, B.; de Albuquerque, L.G. Phenotypic relationship of female sexual precocity with production and reproduction traits in beef cattle using multivariate statistical techniques. Ital. J. Anim. Sci. 2019, 18, 182–188. [Google Scholar] [CrossRef] [Green Version]
- Boligon, A.A.; De Albuquerque, L.G.; Mercadante, M.E.Z.; Lôbo, R.B. Study of relations among age at first calving, average weight gains and weights from weaning to maturity in Nellore cattle. Rev. Bras. Zootec. 2010, 39, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Boligon, A.A.; Albuquerque, L.G. Genetic parameters and relationships of heifer pregnancy and age at first calving with weight gain, yearling and mature weight in Nelore cattle. Livest. Sci. 2011, 141, 12–16. [Google Scholar] [CrossRef] [Green Version]
- Fortes, M.R.S.; Lehnert, S.A.; Bolormaa, S.; Reich, C.; Fordyce, G.; Corbet, N.J.; Whan, V.; Hawken, R.J.; Reverter, A. Finding genes for economically important traits: Brahman cattle puberty. Anim. Prod. Sci. 2012, 52, 143–150. [Google Scholar] [CrossRef]
- Johnston, D.J.; Barwick, S.A.; Corbet, N.J.; Fordyce, G.; Holroyd, R.G.; Williams, P.J.; Burrow, H.M. Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer- and steer-production traits. Anim. Prod. Sci. 2009, 49, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Corbet, N.J.; Allen, J.M.; Laing, A.R.; Fordyce, G.; McGowan, M.R.; Burns, B.M. Using ultrasound to derive new reproductive traits in tropical beef breeds: Implications for genetic evaluation. Anim. Prod. Sci. 2018, 58, 1735–1742. [Google Scholar] [CrossRef]
- Hayes, B.J.; Corbet, N.J.; Allen, J.M.; Laing, A.R.; Fordyce, G.; Lyons, R.; McGowan, M.R.; Burns, B.M. Towards multi-breed genomic evaluations for female fertility of tropical beef cattle. J. Anim. Sci. 2019, 97, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Day, M.L.; Nogueira, G.P. Management of age at puberty in beef heifers to optimize efficiency of beef production. Anim. Front. 2013, 3, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Sartori, R.; Bastos, M.; Baruselli, P.; Gimenes, L.; Ereno, R.; Barros, C. Physiological differences and implications to reproductive management of Bos taurus and Bos indicus cattle in a tropical environment. In Reproduction in Domestic Ruminants VII; Lucy, M., Pate, J., Smith, M., Spencer, T., Eds.; Nottingham University Press: Nottingham, UK, 2010; pp. 357–376. [Google Scholar]
- Costa, E.V.; Ventura, H.T.; Veroneze, R.; Silva, F.F.; Pereira, M.A.; Lopes, P.S. Estimated genetic associations among reproductive traits in Nellore cattle using Bayesian analysis. Anim. Reprod. Sci. 2020, 214, 106305. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, G. Puberty in South American Bos indicus (Zebu) cattle. Anim. Reprod. Sci. 2004, 82–83, 361–372. [Google Scholar] [CrossRef]
- Patterson, D.J.; Perry, R.C.; Kiracofe, G.H.; Bellows, R.A.; Staigmiller, R.B.; Corah, L.R. Management considerations in heifer development and puberty. J. Anim. Sci. 1992, 70, 4018–4035. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, M.V.C.; Pires, A.V.; Santos, M.H.; Silva, R.G.; Oliveira, G.B.; Polizel, D.M.; Biehl, M.V.; Sartori, R.; Nogueira, G.P. A combination of nutrition and genetics is able to reduce age at puberty in Nelore heifers to below 18 months. Animal 2018, 12, 569–574. [Google Scholar] [CrossRef] [Green Version]
- Brumatti, R.C.; Ferraz, J.B.S.; Eler, J.P.; Formigonni, E.I.B. Desenvolvimento de índice de seleção em gado corte sob o enfoque de um modelo bioeconômico*. Arch. Zootec. 2011, 60, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Forni, S.; Albuquerque, L.G. Estimates of genetic correlations between days to calving and reproductive and weight traits in Nelore cattle. J. Anim. Sci. 2005, 83, 1511–1515. [Google Scholar] [CrossRef] [Green Version]
- Bormann, J.M.; Totir, L.R.; Kachman, S.D.; Fernando, R.L.; Wilson, D.E. Pregnancy rate and first-service conception rate in Angus heifers. J. Anim. Sci. 2006, 84, 2022–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigoletto, L.; Santana, M.H.A.; Bressan, F.F.; Eler, J.P.; Nogueira, M.F.G.; Kadarmideen, H.N.; Baruselli, P.S.; Ferraz, J.B.S.; Brito, L.F. Genetic Parameters and Genome-Wide Association Studies for Anti-Müllerian Hormone Levels and Antral Follicle Populations Measured After Estrus Synchronization in Nellore Cattle. Animals 2020, 10, 1185. [Google Scholar] [CrossRef] [PubMed]
- Hazel, L.N.; Dickerson, G.E.; Freeman, A.E. The Selection Index—Then, Now, and for the Future. J. Dairy Sci. 1994, 77, 3236–3251. [Google Scholar] [CrossRef]
- Terakado, A.P.N.; Boligon, A.A.; Baldi, F.; Silva, J.A., IIV; Albuquerque, L.G. Genetic associations between scrotal circumference and female reproductive traits in Nelore cattle1. J. Anim. Sci. 2015, 93, 2706–2713. [Google Scholar] [CrossRef]
- Ponzoni, R.W.; Newman, S. Developing breeding objectives for australian beef cattle production. Anim. Sci. 1989, 49, 35–47. [Google Scholar] [CrossRef]
- Van Eenennaam, A.L.; Weigel, K.A.; Young, A.E.; Cleveland, M.A.; Dekkers, J.C.M. Applied animal genomics: Results from the field. Annu. Rev. Anim. Biosci. 2014, 2, 105–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, B.J.; Lewin, H.A.; Goddard, M.E. The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 2013, 29, 206–214. [Google Scholar] [CrossRef]
- Vargas, C.A.; Elzo, M.A.; Chase, C.C.; Chenoweth, P.J.; Olson, T.A. Estimation of genetic parameters for scrotal circumference, age at puberty in heifers, and hip height in Brahman cattle. J. Anim. Sci. 1998, 76, 2536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossi, D.A.; Venturini, G.C.; Paz, C.C.P.; Bezerra, L.A.F.; Lôbo, R.B.; Oliveira, J.A.; Munari, D.P. Genetic associations between age at first calving and heifer body weight and scrotal circumference in Nelore cattle. J. Anim. Breed. Genet. 2009, 126, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.R.; Barcellos, J.O.J.; Sessim, A.G.; Tarouco, J.U.; Feijó, F.D.; Braccini Neto, J.; Prates, Ê.R.; Canozzi, M.E.A. Relationship of post-weaning growth and age at puberty in crossbred beef heifers. Rev. Bras. Zootec. 2017, 46, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Wolcott, M.L.; Johnston, D.J. The impact of genetic markers for tenderness on steer carcass and feedlot exit and heifer puberty traits in brahman cattle. In Proceedings of the 18th Conference of the Association for the Advancement of Animal Breeding and Genetics, Barossa Valley, Australia, 28 September–1 October 2009; pp. 159–162. [Google Scholar]
- Perry, G.A. Factors affecting puberty in replacement beef heifers. Theriogenology 2016, 86, 373–378. [Google Scholar] [CrossRef]
- Pereira, E.; Oliveira, H.N.; Eler, J.P.; Silva, J.A.; Van Melis, M.H. Comparison among three approaches for evaluation of sexual precocity in Nellore cattle. Animal 2007, 1, 411–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunes, L.C.; Baldi, F.; e Costa, M.F.O.; Lobo, R.B.; Lopes, F.B.; Magnabosco, C.U. Genetic-quantitative analysis for reproductive traits in Nellor cattle selected for sexual precocity. Anim. Prod. Sci. 2020, 60, 896. [Google Scholar] [CrossRef]
- Irano, N.; De Camargo, G.M.F.; Costa, R.B.; Terakado, A.P.N.; Magalhães, A.F.B.; Silva, R.M.D.O.; Dias, M.M.; Bignardi, A.B.; Baldi, F.; Carvalheiro, R.; et al. Genome-wide association study for indicator traits of sexual precocity in Nellore cattle. PLoS ONE 2016, 11, e0159502. [Google Scholar] [CrossRef]
- Boligon, A.A.; Rorato, P.R.N.; de Albuquerque, L.G. Correlações genéticas entre medidas de perímetro escrotal e características produtivas e reprodutivas de fêmeas da raça Nelore. Rev. Bras. Zootec. 2007, 36, 565–571. [Google Scholar] [CrossRef] [Green Version]
- Meirelles, S.L.; Espasandin, A.C.; Mattar, M.; de Queiroz, S.A. Genetic and environmental effects on sexual precocity traits in Nellore cattle. Rev. Bras. Zootec. 2009, 38, 1488–1493. [Google Scholar] [CrossRef] [Green Version]
- Moreira, H.L.; Buzanskas, M.E.; Munari, D.P.; Canova, É.B.; Lôbo, R.B.; de Paz, C.C.P. Reproductive traits selection in Nelore beef cattle. Ciência Agrotecnol. 2015, 39, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Abreu Silva, B.C.; Eler, J.P.; Santana, M.L.; Mattos, E.C.; Menezes, I.R.; Ferraz, J.B.S. Genetic association between mature weight and early growth and heifer pregnancy traits in Nellore cattle. Livest. Sci. 2018, 211, 61–65. [Google Scholar] [CrossRef]
- Yokoo, M.J.; Lôbo, R.B.; Magnabosco, C.U.; Rosa, G.J.M.; Forni, S.; Sainz, R.D.; Albuquerque, L.G. Genetic correlation of traits measured by ultrasound at yearling and 18 months of age in Nellore beef cattle. Livest. Sci. 2015, 180, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Lacerda, V.V.; Campos, G.S.; Roso, V.M.; Souza, F.R.P.; Brauner, C.C.; Boligon, A.A. Effect of mature size and body condition of Nelore females on the reproductive performance. Theriogenology 2018, 118, 27–33. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.R.; Ventura, H.T.; Costa, E.V.; Pereira, M.A.; Veroneze, R.; Duarte, M.D.S.; Dias de Siqueira, O.H.G.B.; Fonseca e Silva, F. Meta-analysis of genetic-parameter estimates for reproduction, growth and carcass traits in Nellore cattle by using a random-effects model. Anim. Prod. Sci. 2018, 58, 1575. [Google Scholar] [CrossRef]
- Kluska, S.; Olivieri, B.F.; Bonamy, M.; Chiaia, H.L.J.; Feitosa, F.L.B.; Berton, M.P.; Peripolli, E.; Lemos, M.V.A.; Tonussi, R.L.; Lôbo, R.B.; et al. Estimates of genetic parameters for growth, reproductive, and carcass traits in Nelore cattle using the single step genomic BLUP procedure. Livest. Sci. 2018, 216, 203–209. [Google Scholar] [CrossRef]
- Silva, M.R.; Pedrosa, V.B.; Silva, J.C.B.; Eler, J.P.; Guimarães, J.D.; Albuquerque, L.G. Testicular traits as selection criteria for young Nellore bulls. J. Anim. Sci. 2011, 89, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, P.A.; Grossi, D.A.; Savegnago, R.P.; Buzanskas, M.E.; Urbinati, I.; Bezerra, L.A.F.; Lôbo, R.B.; Munari, D.P. Estimates of genetic parameters and genetic trends for reproductive traits and weaning weight in Tabapuã cattle1,2. J. Anim. Sci. 2015, 93, 5175–5185. [Google Scholar] [CrossRef]
- Cavani, L.; Garcia, D.A.; Carreño, L.O.D.; Ono, R.K.; Pires, M.P.; Farah, M.M.; Ventura, H.T.; Millen, D.D.; Fonseca, R. Estimates of genetic parameters for reproductive traits in Brahman cattle breed1. J. Anim. Sci. 2015, 93, 3287–3291. [Google Scholar] [CrossRef] [Green Version]
- Paterno, F.M.; Buzanskas, M.E.; Koury Filho, W.; Lôbo, R.B.; Queiroz, S.A. Genetic analysis of visual assessment and body weight traits and their relationships with reproductive traits in Nellore cattle. J. Agric. Sci. 2017, 155, 679–687. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, P.I.; Campos, G.S.; Lôbo, R.B.; Souza, F.R.P.; Brauner, C.C.; Boligon, A.A. Genetic analysis of age at first calving, accumulated productivity, stayability and mature weight of Nellore females. Theriogenology 2018, 108, 81–87. [Google Scholar] [CrossRef]
- Costa, R.B.; Camargo, G.M.; Diaz, I.D.; Irano, N.; Dias, M.M.; Carvalheiro, R.; Boligon, A.A.; Baldi, F.; Oliveira, H.N.; Tonhati, H.; et al. Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. Genet. Sel. Evol. 2015, 47, 67. [Google Scholar] [CrossRef] [Green Version]
- Eler, J.P.; Bignardi, A.B.; Ferraz, J.B.S.; Santana, M.L. Genetic relationships among traits related to reproduction and growth of Nelore females. Theriogenology 2014, 82, 708–714. [Google Scholar] [CrossRef]
- Valente, T.S.; Sant’Anna, A.C.; Baldi, F.; Albuquerque, L.G.; da Costa, M.J.R.P. Genetic association between temperament and sexual precocity indicator traits in Nellore cattle. J. Appl. Genet. 2015, 56, 349–354. [Google Scholar] [CrossRef]
- Van Melis, M.H.; Eler, J.P.; Rosa, G.J.M.; Ferraz, J.B.S.; Figueiredo, L.G.G.; Mattos, E.C.; Oliveira, H.N. Additive genetic relationships between scrotal circumference, heifer pregnancy, and stayability in Nellore cattle. J. Anim. Sci. 2010, 88, 3809–3813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonamy, M.; Kluska, S.; Peripolli, E.; de Lemos, M.V.A.; Amorim, S.T.; Vaca, R.J.; Lôbo, R.B.; de Castro, L.M.; de Faria, C.U.; Borba Ferrari, F.; et al. Genetic association between different criteria to define sexual precocious heifers with growth, carcass, reproductive and feed efficiency indicator traits in Nellore cattle using genomic information. J. Anim. Breed. Genet. 2019, 136, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.I.; Ferreira, I.A.; Silveira, D.D.; Campos, G.S.; Souza, F.R.P.; Carvalheiro, R.; Boligon, A.A. Reproductive performance of cows and genetic correlation with weight gains and principal components of traits used in selection of Nelore cattle. Livest. Sci. 2019, 229, 77–84. [Google Scholar] [CrossRef]
- Eler, J.P.; Silva, J.A., IIV; Ferraz, J.B.S.; Dias, F.; Oliveira, H.N.; Evans, J.L.; Golden, B.L. Genetic evaluation of the probability of pregnancy at 14 months for Nellore heifers1. J. Anim. Sci. 2002, 80, 951–954. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.A. A review of relationships between aspects of reproduction in beef heifers and their lifetime production: 1. Associations with fertility in the first joining season and with age at first joining. Anim. Breed. Abstr. 1980, 48, 655–675. [Google Scholar]
- Formigoni, I.B.; Ferraz, J.B.S.; Silva, J.A.I.V.; Eler, J.P.; Brumatti, R.C. Valores econômicos para habilidade de permanência e probabilidade de prenhez aos 14 meses em bovinos de corte. Arq. Bras. Med. Veterinária Zootec. 2005, 57, 220–226. [Google Scholar] [CrossRef]
- Boligon, A.A.; Ayres, D.R.; Pereira, R.J.; Morotti, N.P.; Albuquerque, L.G. Genetic associations of visual scores with subsequent rebreeding and days to first calving in Nellore cattle. J. Anim. Breed. Genet. 2012, 129, 448–456. [Google Scholar] [CrossRef]
- Van Melis, M.H.; Figueiredo, L.G.G.; Oliveira, H.N.; Eler, J.P.; Rosa, G.J.M.; Santana, M.L., Jr.; Rezende, F.M.; Ferraz, J.B.S. Quantitative genetic study of age at subsequent rebreeding in Nellore cattle by using survival analysis. Genet. Mol. Res. 2014, 13, 4071–4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vasconcelos Silva, J.A.I.; Bignardi, A.B.; Eler, J.P.; Ferraz, J.B.S.; Mercadante, M.E.Z. Análises alternativas no estudo da reconcepção de novilhas Nelore. Bol. Indústria Anim. 2008, 65, 131–135. [Google Scholar]
- Boligon, A.A.; Baldi, F.; Albuquerque, L.G. Genetic correlations between heifer subsequent rebreeding and age at first calving and growth traits in Nellore cattle by Bayesian inference. Genet. Mol. Res. 2012, 11, 4516–4524. [Google Scholar] [CrossRef]
- Schatz, T.J.; Jayawardhana, G.A.; Golding, R.; Hearnden, M.N. Selection for fertility traits in Brahmans increases heifer pregnancy rates from yearling mating. Anim. Prod. Sci. 2010, 50, 345. [Google Scholar] [CrossRef]
- Eler, J.; Santana Junior, M.; Ferraz, J.B. Seleção para precocidade sexual e produtividade da fêmea em bovinos de corte. Estudos 2010, 37, 699–711. [Google Scholar]
- Teixeira, D.B.A.; Fernandes, G.A.; Dos Santos Silva, D.B.; Costa, R.B.; Takada, L.; Gordo, D.G.M.; Bresolin, T.; Carvalheiro, R.; Baldi, F.; De Albuquerque, L.G. Genomic analysis of stayability in Nellore cattle. PLoS ONE 2017, 12, e0179076. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.F.A.; Dórea, J.R.R.; de Magalhães Rosa, G.J. Image Analysis and Computer Vision Applications in Animal Sciences: An Overview. Front. Vet. Sci. 2020, 7, 551269. [Google Scholar] [CrossRef]
- Martins, B.M.; Mendes, A.L.C.; Silva, L.F.; Moreira, T.R.; Costa, J.H.C.; Rotta, P.P.; Chizzotti, M.L.; Marcondes, M.I. Estimating body weight, body condition score, and type traits in dairy cows using three dimensional cameras and manual body measurements. Livest. Sci. 2020, 236, 104054. [Google Scholar] [CrossRef]
- Cominotte, A.; Fernandes, A.F.A.; Dorea, J.R.R.; Rosa, G.J.M.; Ladeira, M.M.; van Cleef, E.H.C.B.; Pereira, G.L.; Baldassini, W.A.; Machado Neto, O.R. Automated computer vision system to predict body weight and average daily gain in beef cattle during growing and finishing phases. Livest. Sci. 2020, 232, 103904. [Google Scholar] [CrossRef]
- Bercovich, A.; Edan, Y.; Alchanatis, V.; Moallem, U.; Parmet, Y.; Honig, H.; Maltz, E.; Antler, A.; Halachmi, I. Development of an automatic cow body condition scoring using body shape signature and Fourier descriptors. J. Dairy Sci. 2013, 96, 8047–8059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, F.M.; Mercadante, M.E.Z.; Barros, C.M.; Satrapa, R.A.; Silva, J.A.V.; Oliveira, L.Z.; Saraiva, N.Z.; Oliveira, C.S.; Garcia, J.M. Reproductive tract development and puberty in two lines of Nellore heifers selected for postweaning weight. Theriogenology 2013, 80, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Talukder, S.; Kerrisk, K.L.; Ingenhoff, L.; Thomson, P.C.; Garcia, S.C.; Celi, P. Infrared technology for estrus detection and as a predictor of time of ovulation in dairy cows in a pasture-based system. Theriogenology 2014, 81, 925–935. [Google Scholar] [CrossRef]
- Oliveira, H.; Lôbo, R.; Pereira, C. Relationships among growth curve parameters, weights and reproductives traits in Guzera beef cows. In Proceedings of the 5th World Congress on Genetics applied to Livestock Production, Guelph, ON, Canada, 7–12 August 1994; pp. 189–192. [Google Scholar]
- Gaviolli, V.R.N.; Buzanskas, M.E.; Cruz, V.A.R.; Savegnago, R.P.; Munari, D.P.; Freitas, A.R.; Alencar, M.M. Genetic associations between weight at maturity and maturation rate with ages and weights at first and second calving in Canchim beef cattle. J. Appl. Genet. 2012, 53, 331–335. [Google Scholar] [CrossRef]
- Inoue, K.; Hosono, M.; Oyama, H.; Hirooka, H. Genetic associations between reproductive traits for first calving and growth curve characteristics of Japanese Black cattle. Anim. Sci. J. 2020, 91, e13467. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.; Goddard, M. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 2010, 185, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Kemper, K.; Savin, K.; Hayes, B.J.; Barendse, W.; Zhang, Y.; Reich, C.M.; Mason, B.A.; Bunch, R.J.; et al. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J. Anim. Sci. 2013, 91, 3088–3104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mota, R.R.; e Silva, F.F.; Guimarães, S.E.F.; Hayes, B.; Fortes, M.R.S.; Kelly, M.J.; Guimarães, J.D.; Penitente-Filho, J.M.; Ventura, H.T.; Moore, S. Benchmarking Bayesian genome enabled-prediction models for age at first calving in Nellore cows. Livest. Sci. 2018, 211, 75–79. [Google Scholar] [CrossRef]
- Alves, A.A.C.; Espigolan, R.; Bresolin, T.; Costa, R.M.; Fernandes Júnior, G.A.; Ventura, R.V.; Carvalheiro, R.; Albuquerque, L.G. Genome-enabled prediction of reproductive traits in Nellore cattle using parametric models and machine learning methods. Anim. Genet. 2021, 52, 32–46. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Fernandes, G.A.; Herrera, A.C.; Scalez, D.C.B.; Espigolan, R.; Magalhães, A.F.B.; Carvalheiro, R.; Baldi, F.; Albuquerque, L.G. Genomic reaction norm models exploiting genotype × environment interaction on sexual precocity indicator traits in Nellore cattle. Anim. Genet. 2020, 51, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Warburton, C.L.; Engle, B.N.; Ross, E.M.; Costilla, R.; Moore, S.S.; Corbet, N.J.; Allen, J.M.; Laing, A.R.; Fordyce, G.; Lyons, R.E.; et al. Use of whole-genome sequence data and novel genomic selection strategies to improve selection for age at puberty in tropically-adapted beef heifers. Genet. Sel. Evol. 2020, 52, 28. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 218–223. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Wiggans, G.R. Derivation, Calculation, and Use of National Animal Model Information. J. Dairy Sci. 1991, 74, 2737–2746. [Google Scholar] [CrossRef]
- Garrick, D.J. The nature, scope and impact of genomic prediction in beef cattle in the United States. Genet. Sel. Evol. 2011, 43, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Lopes, F.B.; Fernandes Júnior, G.A.; Rosa, G.J.M.; Magalhães, A.F.B.; Carvalheiro, R.; Albuquerque, L.G. Genome-wide scan highlights the role of candidate genes on phenotypic plasticity for age at first calving in Nellore heifers. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kolmodin, R.; Strandberg, E.; Jorjani, H.; Danell, B. Selection in the presence of a genotype by environment interaction: Response in environmental sensitivity. Anim. Sci. 2003, 76, 375–385. [Google Scholar] [CrossRef]
- Falconer, D.S. Selection in different environments: Effects on environmental sensitivity (reaction norm) and on mean performance. Genet. Res. 1990, 56, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Carvalheiro, R.; Costilla, R.; Neves, H.H.R.; Albuquerque, L.G.; Moore, S.; Hayes, B.J. Unraveling genetic sensitivity of beef cattle to environmental variation under tropical conditions. Genet. Sel. Evol. 2019, 51, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Mota, L.F.M.; Costa, L.S.; Garzón, N.A.M.; Passafaro, T.L.; Silva, D.O.; Abreu, L.R.A.; Verardo, L.L.; Bonafé, C.M.; Ventura, H.T. Unraveling the effect of body structure score on phenotypic plasticity for body weight at different ages in Guzerat cattle. Livest. Sci. 2019, 229, 98–104. [Google Scholar] [CrossRef]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [Green Version]
- Santana, M.L., Jr.; Eler, J.P.; Bignardi, A.B.; Boligon, A.A.; Ferraz, J.B.S. Genetic correlation between growth and reproductive performance of beef females depends on environment. Anim. Prod. Sci. 2018, 58, 1201. [Google Scholar] [CrossRef] [Green Version]
- Samadi, F.; Blache, D.; Martin, G.B.; D’Occhio, M.J. Nutrition, metabolic profiles and puberty in Brahman (Bos indicus) beef heifers. Anim. Reprod. Sci. 2014, 146, 134–142. [Google Scholar] [CrossRef]
- Chiaia, H.L.J.; De Lemos, M.V.A.; Venturini, G.C.; Aboujaoude, C.; Berton, M.P.; Feitosa, F.B.; Carvalheiro, R.; Albuquerque, L.G.; De Oliveira, H.N.; Baldi, F. Genotype × environment interaction for age at first calving, scrotal circumference, and yearling weight in Nellore cattle using reaction norms in multitrait random regression models. J. Anim. Sci. 2015, 93, 1503–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santana, M.L.; Eler, J.P.; Oliveira, G.A.; Bignardi, A.B.; Pereira, R.J.; Ferraz, J.B.S. Genetic variation in Nelore heifer pregnancy due to heat stress during the breeding season. Livest. Sci. 2018, 218, 101–107. [Google Scholar] [CrossRef]
- Mulder, H.A.; Bijma, P.; Hill, W.G. Selection for uniformity in livestock by exploiting genetic heterogeneity of residual variance. Genet. Sel. Evol. 2008, 40, 37. [Google Scholar] [CrossRef] [Green Version]
- Félix, M.-A.; Barkoulas, M. Pervasive robustness in biological systems. Nat. Rev. Genet. 2015, 16, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I. Breeding and Genetics Symposium: Resilience and lessons from studies in genetics of heat stress1,2. J. Anim. Sci. 2017, 95, 1780–1787. [Google Scholar] [CrossRef] [Green Version]
- Hermesch, S.; Li, L.; Doeschl-Wilson, A.B.; Gilbert, H. Selection for productivity and robustness traits in pigs. Anim. Prod. Sci. 2015, 55, 1437. [Google Scholar] [CrossRef]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2019, 9, 692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knap, P.W. Breeding robust pigs. Aust. J. Exp. Agric. 2005, 45, 763. [Google Scholar] [CrossRef]
- Mulder, H.A.; Bijma, P. Effects of genotype × environment interaction on genetic gain in breeding programs. J. Anim. Sci. 2005, 83, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Mulder, H.A. Genomic Selection Improves Response to Selection in Resilience by Exploiting Genotype by Environment Interactions. Front. Genet. 2016, 7, 1–11. [Google Scholar] [CrossRef]
- Nirea, K.G.; Meuwissen, T.H.E. Improving production efficiency in the presence of genotype by environment interactions in pig genomic selection breeding programmes. J. Anim. Breed. Genet. 2017, 134, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Mota, R.R.; Lopes, P.S.; Tempelman, R.J.; Silva, F.F.; Aguilar, I.; Gomes, C.C.G.; Cardoso, F.F. Genome-enabled prediction for tick resistance in Hereford and Braford beef cattle via reaction norm models. J. Anim. Sci. 2016, 94, 1834–1843. [Google Scholar] [CrossRef] [Green Version]
- Silva, F.F.; Mulder, H.A.; Knol, E.F.; Lopes, M.S.; Guimarães, S.E.F.; Lopes, P.S.; Mathur, P.K.; Viana, J.M.S.; Bastiaansen, J.W.M. Sire evaluation for total number born in pigs using a genomic reaction norms approach1. J. Anim. Sci. 2014, 92, 3825–3834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, B.J.; Daetwyler, H.D.; Goddard, M.E. Models for Genome × Environment Interaction: Examples in Livestock. Crop Sci. 2016, 56, 1–9. [Google Scholar] [CrossRef]
- Miszura, A.A.; Ferraz, M.V.C.; Cardoso, R.C.; Polizel, D.M.; Oliveira, G.B.; Barroso, J.P.R.; Gobato, L.G.M.; Nogueira, G.P.; Biava, J.S.; Ferreira, E.M.; et al. Implications of growth rates and compensatory growth on puberty attainment in Nellore heifers. Domest. Anim. Endocrinol. 2021, 74, 106526. [Google Scholar] [CrossRef] [PubMed]
- D’Occhio, M.J.; Baruselli, P.S.; Campanile, G. Influence of nutrition, body condition, and metabolic status on reproduction in female beef cattle: A review. Theriogenology 2019, 125, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, C.J.; Toma, L.M.; Hunter, M.G. Nutritional effects on oocyte and embryo development in mammals: Implications for reproductive efficiency and environmental sustainability. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Reecy, J.M. Animal QTLdb: Beyond a repository-A public platform for QTL comparisons and integration with diverse types of structural genomic information. Mamm. Genome 2007, 18, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Fortes, M.R.S.; Snelling, W.M.; Reverter, A.; Nagaraj, S.H.; Lehnert, S.A.; Hawken, R.J.; DeAtley, K.L.; Peters, S.O.; Silver, G.A.; Rincon, G.; et al. Gene network analyses of first service conception in Brangus heifers: Use of genome and trait associations, hypothalamic-transcriptome information, and transcription factors1. J. Anim. Sci. 2012, 90, 2894–2906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawken, R.J.; Zhang, Y.D.; Fortes, M.R.S.; Collis, E.; Barris, W.C.; Corbet, N.J.; Williams, P.J.; Fordyce, G.; Holroyd, R.G.; Walkley, J.R.W.; et al. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J. Anim. Sci. 2012, 90, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Fortes, M.R.S.; Kemper, K.; Sasazaki, S.; Reverter, A.; Pryce, J.E.; Barendse, W.; Bunch, R.; McCulloch, R.; Harrison, B.; Bolormaa, S.; et al. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian Beef cattle. Anim. Genet. 2013, 44, 636–647. [Google Scholar] [CrossRef]
- Peters, S.O.; Kizilkaya, K.; Garrick, D.J.; Fernando, R.L.; Reecy, J.M.; Weaber, R.L.; Silver, G.A.; Thomas, M.G. Heritability and bayesian genome-wide association study of first service conception and pregnancy in Brangus heifers. J. Anim. Sci. 2013, 91, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.V.; Matos, M.C.; Seno, L.O.; Romero, A.R.S.; Garcia, J.F.; Grisolia, A.B. Genome wide association study on early puberty in Bos indicus. Genet. Mol. Res. 2016, 15, 1–6. [Google Scholar] [CrossRef]
- Buzanskas, M.E.; do Amaral Grossi, D.; Ventura, R.V.; Schenkel, F.S.; Chud, T.C.S.; Stafuzza, N.B.; Rola, L.D.; Meirelles, S.L.C.; Mokry, F.B.; Munari, D.P.; et al. Candidate genes for male and female reproductive traits in Canchim beef cattle. J. Anim. Sci. Biotechnol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mota, R.R.; Guimarães, S.E.F.; Fortes, M.R.S.; Hayes, B.; Silva, F.F.; Verardo, L.L.; Kelly, M.J.; Campos, C.F.; Guimarães, J.D.; Wenceslau, R.R.; et al. Genome-wide association study and annotating candidate gene networks affecting age at first calving in Nellore cattle. J. Anim. Breed Genet. 2017, 134, 484–492. [Google Scholar] [CrossRef] [Green Version]
- Oliveira Júnior, G.A.; Perez, B.C.; Cole, J.B.; Santana, M.H.A.; Silveira, J.; Mazzoni, G.; Ventura, R.V.; Santana, M.L.; Kadarmideen, H.N.; Garrick, D.J.; et al. Genomic study and medical subject headings enrichment analysis of early pregnancy rate and antral follicle numbers in Nelore heifers. J. Anim. Sci. 2017, 95, 4796–4812. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Enculescu, C.; Neto, L.R.P.; Lehnert, S.A.; McCulloch, R.; Hayes, B. Candidate mutations used to aid the prediction of genetic merit for female reproductive traits in tropical beef cattle. Rev. Bras. Zootec. 2018, 47, e20170226. [Google Scholar] [CrossRef] [Green Version]
- Melo, T.P.; Fortes, M.R.S.; Bresolin, T.; Mota, L.F.M.; Albuquerque, L.G.; Carvalheiro, R. Multitrait meta-analysis identified genomic regions associated with sexual precocity in tropical beef cattle1. J. Anim. Sci. 2018, 96, 4087–4099. [Google Scholar] [CrossRef] [PubMed]
- Melo, T.P.; Fortes, M.R.S.; Fernandes Junior, G.A.; Albuquerque, L.G.; Carvalheiro, R. RAPID COMMUNICATION: Multi-breed validation study unraveled genomic regions associated with puberty traits segregating across tropically adapted breeds. J. Anim. Sci. 2019, 97, 3027–3033. [Google Scholar] [CrossRef]
- Oliveira Júnior, G.A.; Santos, D.J.A.; Cesar, A.S.M.; Boison, S.A.; Ventura, R.V.; Perez, B.C.; Garcia, J.F.; Ferraz, J.B.S.; Garrick, D.J. Fine mapping of genomic regions associated with female fertility in Nellore beef cattle based on sequence variants from segregating sires. J. Anim. Sci. Biotechnol. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fortes, M.R.S.; Reverter, A.; Zhang, Y.; Collis, E.; Nagaraj, S.H.; Jonsson, N.N.; Prayaga, K.C.; Barris, W.; Hawken, R.J. Association weight matrix for the genetic dissection of puberty in beef cattle. Proc. Natl. Acad. Sci. USA 2010, 107, 13642–13647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortes, M.R.S.; Nguyen, L.T.; Weller, M.M.D.C.A.; Cánovas, A.; Islas-Trejo, A.; Porto-Neto, L.R.; Reverter, A.; Lehnert, S.A.; Boe-Hansen, G.B.; Thomas, M.G.; et al. Transcriptome analyses identify five transcription factors differentially expressed in the hypothalamus of post- versus prepubertal Brahman heifers. J. Anim. Sci. 2016, 94, 3693–3702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, L.; Barbero, M.M.D.; Oliveira, H.N.; De Camargo, G.M.F.; Fernandes, G.A.; Aspilcueta-Borquis, R.R.; Souza, F.R.P.; Boligon, A.A.; Melo, T.P.; Regatieri, I.C.; et al. Genomic association for sexual precocity in beef heifers using pre-selection of genes and haplotype reconstruction. PLoS ONE 2018, 13, e0190197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giguère, V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008, 29, 677–696. [Google Scholar] [CrossRef] [Green Version]
- Kubo, M.; Ijichi, N.; Ikeda, K.; Horie-Inoue, K.; Takeda, S.; Inoue, S. Modulation of adipogenesis-related gene expression by estrogen-related receptor γ during adipocytic differentiation. Biochim. Biophys. Acta-Gene Regul. Mech. 2009, 1789, 71–77. [Google Scholar] [CrossRef]
- Villena, J.A.; Hock, M.B.; Chang, W.Y.; Barcas, J.E.; Giguère, V.; Kralli, A. Orphan nuclear receptor estrogen-related receptor α is essential for adaptive themogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 1418–1423. [Google Scholar] [CrossRef] [Green Version]
- Reverter, A.; Porto-Neto, L.R.; Fortes, M.R.S.; Kasarapu, P.; de Cara, M.A.R.; Burrow, H.M.; Lehnert, S.A. Genomic inbreeding depression for climatic adaptation of tropical beef cattle. J. Anim. Sci. 2017, 95, 3809. [Google Scholar] [CrossRef] [PubMed]
- Porto-Neto, L.R.; Reverter, A.; Prayaga, K.C.; Chan, E.K.F.; Johnston, D.J.; Hawken, R.J.; Fordyce, G.; Garcia, J.F.; Sonstegard, T.S.; Bolormaa, S.; et al. The genetic architecture of climatic adaptation of tropical cattle. PLoS ONE 2014, 9, e113284. [Google Scholar] [CrossRef] [Green Version]
- Medeiros De Oliveira Silva, R.; Stafuzza, N.B.; Fragomeni, B.D.O.; Ferreira De Camargo, G.M.; Ceacero, T.M.; Cyrillo, J.N.D.S.G.; Baldi, F.; Boligon, A.A.; Mercadante, M.E.Z.; Lourenco, D.L.; et al. Genome-wide association study for carcass traits in an experimental nelore cattle population. PLoS ONE 2017, 12, e0169860. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Kuehn, L.A.; Smith, T.P.L.; Ferrell, C.L.; Jenkins, T.G.; Freetly, H.C.; Snelling, W.M. A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle. Anim. Genet. 2012, 43, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Saatchi, M.; Schnabel, R.D.; Taylor, J.F.; Garrick, D.J. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genom. 2014, 15, 442. [Google Scholar] [CrossRef] [Green Version]
- Fernandes Júnior, G.A.; Rosa, G.J.M.; Costa, R.B.; Carvalheiro, R.; Chardulo, L.A.L.; Gordo, D.G.M.; Baldi, F.; Oliveira, H.N.; Silva, R.M.O.; Tonhati, H.; et al. Genome-assisted multiple-trait analysis of carcass traits in Nellore cattle. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Bolormaa, S.; Pryce, J.E.; Reverter, A.; Zhang, Y.; Barendse, W.; Kemper, K.; Tier, B.; Savin, K.; Hayes, B.J.; Goddard, M.E. A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle. PLoS Genet. 2014, 10, e1004198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, F.; Rings, F.; Mamo, S.; Holker, M.; Kuzmany, A.; Besenfelder, U.; Havlicek, V.; Mehta, J.P.; Tesfaye, D.; Schellander, K.; et al. Effect of elevated circulating progesterone concentration on bovine blastocyst development and global transcriptome following endoscopic transfer of in vitro produced embryos to the bovine oviduct. Biol. Reprod. 2010, 83, 707–719. [Google Scholar] [CrossRef]
- Belleannée, C.; Calvo, E.; Thimon, V.; Cyr, D.G.; Légaré, C.; Garneau, L.; Sullivan, R. Role of microRNAs in controlling gene expression in different segments of the human epididymis. PLoS ONE 2012, 7, e34996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, R.; Su, J.; Zheng, L.; Jin, M.; Hou, Y.; Ma, Z.; Guo, T.; Zhu, S.; Ma, X.; Ahmed, E.; et al. Cloning and distribution of neuropeptide W and its receptors in pigs. Res. Vet. Sci. 2015, 101, 106–116. [Google Scholar] [CrossRef]
- Fernandes Júnior, G.A.; Costa, R.B.; de Camargo, G.M.F.; Carvalheiro, R.; Rosa, G.J.M.; Baldi, F.; Garcia, D.A.; Gordo, D.G.M.; Espigolan, R.; Takada, L.; et al. Genome scan for postmortem carcass traits in Nellore cattle. J. Anim. Sci. 2016, 94, 4087–4095. [Google Scholar] [CrossRef]
- Ma, L.; Cole, J.B.; Da, Y.; VanRaden, P.M. Symposium review: Genetics, genome-wide association study, and genetic improvement of dairy fertility traits. J. Dairy Sci. 2019, 102, 3735–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Den Berg, I.; Xiang, R.; Jenko, J.; Pausch, H.; Boussaha, M.; Schrooten, C.; Tribout, T.; Gjuvsland, A.B.; Boichard, D.; Nordbø, Ø.; et al. Meta-analysis for milk fat and protein percentage using imputed sequence variant genotypes in 94,321 cattle from eight cattle breeds. Genet. Sel. Evol. 2020, 52, 1–16. [Google Scholar] [CrossRef]
- Tahir, M.S.; Porto-Neto, L.R.; Gondro, C.; Shittu, O.B.; Wockner, K.; Tan, A.W.L.; Smith, H.R.; Gouveia, G.C.; Kour, J.; Fortes, M.R.S. Meta-Analysis of Heifer Traits Identified Reproductive Pathways in Bos indicus Cattle. Genes 2021, 12, 768. [Google Scholar] [CrossRef] [PubMed]
- Peñagaricano, F.; Weigel, K.A.; Rosa, G.J.M.; Khatib, H. Inferring Quantitative Trait Pathways Associated with Bull Fertility from a Genome-Wide Association Study. Front. Genet. 2013, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- MacLeod, I.M.; Bowman, P.J.; Vander Jagt, C.J.; Haile-Mariam, M.; Kemper, K.E.; Chamberlain, A.J.; Schrooten, C.; Hayes, B.J.; Goddard, M.E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brøndum, R.F.; Liao, X.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef] [PubMed]
- McClure, M.C.; Bickhart, D.; Null, D.; VanRaden, P.; Xu, L.; Wiggans, G.; Liu, G.; Schroeder, S.; Glasscock, J.; Armstrong, J.; et al. Bovine Exome Sequence Analysis and Targeted SNP Genotyping of Recessive Fertility Defects BH1, HH2, and HH3 Reveal a Putative Causative Mutation in SMC2 for HH3. PLoS ONE 2014, 9, e92769. [Google Scholar] [CrossRef] [PubMed]
- Whiston, R.; Finlay, E.K.; McCabe, M.S.; Cormican, P.; Flynn, P.; Cromie, A.; Hansen, P.J.; Lyons, A.; Fair, S.; Lonergan, P.; et al. A dual targeted β-defensin and exome sequencing approach to identify, validate and functionally characterise genes associated with bull fertility. Sci. Rep. 2017, 7, 12287. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Novo, A.; Pérez-Garnelo, S.S.; Villagrá, A.; Pérez-Villalobos, N.; Astiz, S. The Effect of Stress on Reproduction and Reproductive Technologies in Beef Cattle—A Review. Animals 2020, 10, 2096. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.C.; da Silva Andrade, S.C.; de Melo, G.D.; Motta, I.G.; Coutinho, L.L.; Gonella-Diaza, A.M.; Binelli, M.; Pugliesi, G. Early pregnancy-induced transcripts in peripheral blood immune cells in Bos indicus heifers. Sci. Rep. 2020, 10, 13733. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Reverter, A.; Cánovas, A.; Venus, B.; Islas-Trejo, A.; Porto-Neto, L.R.; Lehnert, S.A.; Medrano, J.F.; Moore, S.S.; Fortes, M.R.S. Global differential gene expression in the pituitary gland and the ovaries of pre- and postpubertal Brahman heifers1. J. Anim. Sci. 2017, 95, 599–615. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Zacchi, L.F.; Schulz, B.L.; Moore, S.S.; Fortes, M.R.S. Adipose tissue proteomic analyses to study puberty in Brahman heifers. J. Anim. Sci. 2018, 96, 2392–2398. [Google Scholar] [CrossRef]
- Tahir, M.S.; Nguyen, L.T.; Schulz, B.L.; Boe-Hansen, G.A.; Thomas, M.G.; Moore, S.S.; Lau, L.Y.; Fortes, M.R.S. Proteomics Recapitulates Ovarian Proteins Relevant to Puberty and Fertility in Brahman Heifers (Bos indicus L.). Genes 2019, 10, 923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Occhio, M.J.; Baruselli, P.S.; Campanile, G. Metabolic health, the metabolome and reproduction in female cattle: A review. Ital. J. Anim. Sci. 2019, 18, 858–867. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, S. Interactions of Metabolism, Inflammation, and Reproductive Tract Health in the Postpartum Period in Dairy Cattle. Reprod. Domest. Anim. 2012, 47, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Sundrum, A. Metabolic Disorders in the Transition Period Indicate that the Dairy Cows’ Ability to Adapt is Overstressed. Animals 2015, 5, 978–1020. [Google Scholar] [CrossRef]
- Cánovas, A.; Reverter, A.; DeAtley, K.L.; Ashley, R.L.; Colgrave, M.L.; Fortes, M.R.S.; Islas-Trejo, A.; Lehnert, S.; Porto-Neto, L.; Rincón, G.; et al. Multi-Tissue Omics Analyses Reveal Molecular Regulatory Networks for Puberty in Composite Beef Cattle. PLoS ONE 2014, 9, e102551. [Google Scholar] [CrossRef] [Green Version]
- de Souza Fonseca, P.A.; Id-Lahoucine, S.; Reverter, A.; Medrano, J.F.; Fortes, M.S.; Casellas, J.; Miglior, F.; Brito, L.; Carvalho, M.R.S.; Schenkel, F.S.; et al. Combining multi-OMICs information to identify key-regulator genes for pleiotropic effect on fertility and production traits in beef cattle. PLoS ONE 2018, 13, e0205295. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Zacchi, L.F.; Nguyen, L.T.; Raidan, F.; Weller, M.M.D.C.A.; Choo, J.J.Y.; Reverter, A.; Rego, J.P.A.; Boe-Hansen, G.B.; Porto-Neto, L.R.; et al. Pre- and post-puberty expression of genes and proteins in the uterus of Bos indicus heifers: The luteal phase effect post-puberty. Anim. Genet. 2018, 49, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Carvalheiro, R. Genomic Selection in Nelore Cattle in Brazil. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Fries, L.A.; Ferraz, J.B.S. Beef cattle genetic programmes in Brazil. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production; Belo Horizonte, Belo Horizonte, MG, Brazil, 13–18 August 2006. [Google Scholar]
- Eler, J.P.; Silva, J.A., IIV; Evans, J.L.; Ferraz, J.B.S.; Dias, F.; Golden, B.L. Additive genetic relationships between heifer pregnancy and scrotal circumference in Nellore cattle1. J. Anim. Sci. 2004, 82, 2519–2527. [Google Scholar] [CrossRef]
- Eler, J.P.; Ferraz, J.B.S.; Balieiro, J.C.C.; Mattos, E.C.; Mourão, G.B. Genetic correlation between heifer pregnancy and scrotal circumference measured at 15 and 18 months of age in Nellore cattle. Genet. Mol. Res. 2006, 5, 569–580. [Google Scholar]
- Shiotsuki, L.; Silva, J.A.V., II; Tonhati, H.; Albuquerque, L.G. Genetic associations of sexual precocity with growth traits and visual scores of conformation, finishing, and muscling in Nelore cattle1. J. Anim. Sci. 2009, 87, 1591–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickson, R.E.; Lopez-Villalobos, N.; Kenyon, P.R.; Ridler, B.J.; Morris, S.T. Profitability of calving heifers at 2 compared with 3 years of age and the effect of incidence of assistance at parturition on profitability. Anim. Prod. Sci. 2010, 50, 354. [Google Scholar] [CrossRef]
- Núñez-Dominguez, R.; Cundiff, L.V.; Dickerson, G.E.; Gregory, K.E.; Koch, R.M. Lifetime production of beef heifers calving first at two vs three years of age1. J. Anim. Sci. 1991, 69, 3467–3479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capper, J.L. The environmental impact of beef production in the United States: 1977 compared with 2007. J. Anim. Sci. 2011, 89, 4249–4261. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Phenotype | BTA | Pos (Mb) a | Breed | Number of Phenotypes/Genotypes | Reference |
|---|---|---|---|---|---|
| First Service Conception | 1 | 135.38 | Brangus | 861/802 | [117] Fortes et al., 2012 |
| 5 | 56.67 | ||||
| 5 | 70.26 | ||||
| 9 | 82.42 | ||||
| 11 | 95.64 | ||||
| Age at first corpus luteum | 14 | 20–33 | Brahman | 837 | [118] Hawken et al., 2012 |
| 5 | 93–96 | Tropical Composite | 860 | ||
| Age at first corpus luteum | 14 | 25.01 | Brahman | 1007 | [119] Fortes et al., 2013 |
| Tropical Composite | 1111 | ||||
| First Service Conception | 3 | 101.0–101.9 | Brangus | 830/796 | [120] Peters et al., 2013 |
| 8 | 25.0–26.9 | ||||
| 15 | 69.0–69.9 | ||||
| 16 | 43.1–43.9 | ||||
| 19 | 49.0–49.9 | ||||
| 24 | 53.0–53.9 | ||||
| 26 | 8.0–8.9 | ||||
| 26 | 16.0–16.9 | ||||
| 27 | 33.0–33.9 | ||||
| 29 | 22.0–22.9 | ||||
| X | 108.1–108.9 | ||||
| Heifer Pregnancy | 2 | 41.0–41.9 | Brangus | 830/796 | [120] Peters et al., 2013 |
| 4 | 4.0–4.9 | ||||
| 8 | 0.3–0.9 | ||||
| 10 | 91.0–91.9 | ||||
| 13 | 83.0–83.9 | ||||
| 20 | 70.0–70.9 | ||||
| Early Pregnancy | 5 | 8.8–10.12 | Nellore | 73,359/1770 | [40] Irano et al., 2016 |
| 5 | 16.06–17.12 | ||||
| 6 | 10.64–11.66 | ||||
| 7 | 3.12–3.85 | ||||
| 7 | 41.28–42.03 | ||||
| 14 | 22.61–23.39 | ||||
| 18 | 4.26–4.91 | ||||
| 21 | 0.01–3.02 | ||||
| 21 | 61.92–62.53 | ||||
| 27 | 0.99–1.57 | ||||
| Early Puberty | 5 | 78.64 | Nellore | 55 | [121] Nascimento et al., 2016 |
| 6 | 59.02 | ||||
| 9 | 8.44 | ||||
| 10 | 33.82 | ||||
| 22 | 10.41 | ||||
| Age at first calving | 4 | 17.46 | Canchim | 267,002/392 | [122] Buzanskas et al., 2017 |
| 4 | 98.31 | ||||
| 27 | 35.19–35.21 | ||||
| Age at first calving | 2 | 6.17–7.17 | Nellore | 762/2992 | [123] Mota et al., 2017 |
| 8 | 106.27–107.27 | ||||
| 9 | 40.97–46.61 | ||||
| 14 | 16.54–17.53 | ||||
| 14 | 20.39–24.67 | ||||
| 14 | 26.20–28.84 | ||||
| 14 | 31.25–36.95 | ||||
| 16 | 43.94–44.93 | ||||
| 16 | 68.23–69.23 | ||||
| 17 | 57.29–58.28 | ||||
| Early Pregnancy | 5 | 72.52–74.46 | Nellore | 2283/2283 | [124] Oliveira Junior et al., 2017 |
| 5 | 76.52–78.48 | ||||
| 5 | 80.63–82.46 | ||||
| 14 | 22.50–24.48 | ||||
| 14 | 28.56–30.48 | ||||
| 18 | 54.51–56.45 | ||||
| Number of antral follicles | 2 | 122.53–124.48 | Nellore | 1099/2283 | [124] Oliveira Junior et al., 2017 |
| 8 | 6.51–8.47 | ||||
| 11 | 69.52–71.47 | ||||
| 14 | 22.50–24.48 | ||||
| 15 | 8.50–10.46 | ||||
| 16 | 70.50–72.45 | ||||
| 22 | 14.50–16.47 | ||||
| Age at first corpus luteum | 7 | 23 | Brahman | 914 | [125] Fortes et al., 2018 |
| 21 | 23 | ||||
| 19 | 49 | Tropical Composite | 798 | ||
| Meta-analysis for fertility traits | 1 | 118.6 | Nellore | AFC 1796/1796 | [126] Melo et al., 2018 |
| 2 | 95.917 | ||||
| 3 | 49.43 | ||||
| 4 | 110.44 | ||||
| 6 | 118.43 | ||||
| 7 | 94.71 | EP (%) 1849/1849 | |||
| 8 | 68.3 | ||||
| 9 | 75.61 | ||||
| 10 | 16.76 | ||||
| 11 | 104.93 | ||||
| 13 | 16.09 | Brahman | AGECL 1007 | ||
| 14 | 24.71 | ||||
| 15 | 9.06 | ||||
| 16 | 1.92 | ||||
| 21 | 11.43 | ||||
| 24 | 2.27 | ||||
| 26 | 23.4 | ||||
| 27 | 31.92 | ||||
| 29 | 9.17 | ||||
| Age at first corpus luteum | 1 | 43.45 | Tropical Composite | 1097 | [127] Melo et al., 2019 |
| 14 | 25.24 | ||||
| 15 | 9.06 | ||||
| 16 | 24.328 | ||||
| 21 | 6.83 | ||||
| 23 | 27.78 | ||||
| 26 | 23.4 | ||||
| 29 | 9.17 | ||||
| Heifer pregnancy | 5 | 70.5–72.7 | Nellore | 1337/1337 | [128] Oliveira Jr., 2019 |
| 14 | 20.7–24.6 | ||||
| Age at first calving | 1 | 22.86–23.03 | Nellore | 185,356/3760 | [91] Mota et al., 2020 |
| 2 | 105.03–105.38 | ||||
| 3 | 21.19–21.22 | ||||
| 3 | 8.34–8.41 | ||||
| 5 | 9.47–10.87 | ||||
| 6 | 19.49–19.54 | ||||
| 14 | 24.82–25.10 | ||||
| 15 | 35.34–35.64 | ||||
| 17 | 49.64–49.83 | ||||
| 18 | 3.08–4.89 | ||||
| 27 | 31.64–31.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes Júnior, G.A.; Silva, D.A.; Mota, L.F.M.; de Melo, T.P.; Fonseca, L.F.S.; Silva, D.B.d.S.; Carvalheiro, R.; Albuquerque, L.G. Sustainable Intensification of Beef Production in the Tropics: The Role of Genetically Improving Sexual Precocity of Heifers. Animals 2022, 12, 174. https://doi.org/10.3390/ani12020174
Fernandes Júnior GA, Silva DA, Mota LFM, de Melo TP, Fonseca LFS, Silva DBdS, Carvalheiro R, Albuquerque LG. Sustainable Intensification of Beef Production in the Tropics: The Role of Genetically Improving Sexual Precocity of Heifers. Animals. 2022; 12(2):174. https://doi.org/10.3390/ani12020174
Chicago/Turabian StyleFernandes Júnior, Gerardo Alves, Delvan Alves Silva, Lucio Flavio Macedo Mota, Thaise Pinto de Melo, Larissa Fernanda Simielli Fonseca, Danielly Beraldo dos Santos Silva, Roberto Carvalheiro, and Lucia Galvão Albuquerque. 2022. "Sustainable Intensification of Beef Production in the Tropics: The Role of Genetically Improving Sexual Precocity of Heifers" Animals 12, no. 2: 174. https://doi.org/10.3390/ani12020174
APA StyleFernandes Júnior, G. A., Silva, D. A., Mota, L. F. M., de Melo, T. P., Fonseca, L. F. S., Silva, D. B. d. S., Carvalheiro, R., & Albuquerque, L. G. (2022). Sustainable Intensification of Beef Production in the Tropics: The Role of Genetically Improving Sexual Precocity of Heifers. Animals, 12(2), 174. https://doi.org/10.3390/ani12020174







