Simple Summary
Yaks are among the largest high-altitude mammals in the world, and they are ideally adapted to the harsh environmental conditions of the plateau regions. Yaks are thus central to the lives of herdsmen and other local populations in these high-altitude areas. Copy number variations (CNVs) are an important cause of genomic variation in livestock and identifying advantageous CNVs can aid in livestock breeding efforts. In this study, an association between CNVs in the MICALL2 and MOGAT2 genes and Ashidan yak growth traits was confirmed, providing a theoretical foundation for Ashidan yak breeding and meat production efforts.
Abstract
Copy number variations (CNVs) are a result of genomic rearrangement affecting DNA regions over 1 kb in length, and can include inversions, translocations, deletions, and duplications. The molecule interacting with CasL-like protein 2 (MICALL2) gene is primarily associated with mitochondrial protein targeting and exhibits predicted stress fiber colocalization. The monoacylglycerol O-acyltransferase 2 (MOGAT2) gene encodes an enzyme responsible for catalyzing diacylglycerol synthesis from 2-monoacylglycerol and fatty acyl-CoA. For this study, blood samples were obtained from 315 yaks, and the body weight, body length, withers height, and chest girth of these animals were measured at 6, 12, 18, and 30 months of age. Genomic DNA was harvested from the collected blood samples, and CNVs in these samples were detected by qPCR. The resultant data were compared using ANOVAs, revealing significant associations between MICALL2 gene CNVs and body weight at 6 months of age (p < 0.05), body length and chest girth at 30 months of age (p < 0.05), and withers height at 18 months of age (p < 0.01) in Ashidan yaks. Similarly, MOGAT2 CNVs were significantly associated with body weight at 6 and 30 months of age (p < 0.05), and with withers height at 18 months of age (p < 0.01) in these Ashidan yaks. MICALL2 and MOGAT2 gene expression was further analyzed in yak tissue samples, revealing that MICALL2 was most highly expressed in the adipose tissue, whereas MOGAT2 was most highly expressed in the lung. These results thus confirmed the relationship between CNVs in the MICALL2 and MOGAT2 genes and Ashidan yak growth traits, providing a valuable gene locus that can be leveraged for future marker-assisted yak breeding efforts.
1. Introduction
Yaks (Bos grunniens) are among the most important livestock animals bred in the Qinghai-Tibet Plateau region of China. Yaks are ideally suited to life in this high-altitude environment, as they exhibit excellent tolerance for cold and food scarcity [1]. Initially established in 2019, Ashidan yaks are the first hornless yak breed. Relative to other breeds, these Ashidan yaks are more docile while remaining strong, with dark coloration. They are also well-suited to increased breeding density in an enclosed setting given their docility and lack of horns. Owing to the harsh climatic conditions of the Qinghai-Tibet Plateau, grazing yaks in this region rely on hay intake for 7 months per year during which the average body weight of these animals drops significantly. However, as Ashidan yaks can be raised in an enclosed environment and provided with supplemental nutrition, their body weight can be maintained at stable levels throughout the year [2]. Studies conducted to date have focused on exploring genetic polymorphisms associated with specific yak traits, thus reflecting the rich genetic diversity of these yak breeds while clarifying the genetic relationships between specific breeds and their wild relatives [3]. In addition to permitting the interrogation of the origins and genetic differentiation of these yak breeds, these studies can provide a robust theoretical foundation for directed breeding efforts, enabling the production of superior animals and animal-derived products through the effective leveraging of these genetic resources.
Copy number variations (CNVs), which include insertions, deletions, and other forms of repetition in genomic DNA sequences over 1 kb in length, are a major source of genetic variation among individuals in a given species. These CNVs can arise due to various forms of genomic rearrangement including translocations, inversions, duplications, or deletions of particular regions of the genome [4]. The resulting genetic polymorphisms often produce distinct phenotypic traits in individuals carrying these CNVs [5]. Since their initial codification as a phenotypically important source of variability in 1936 by the American geneticist Calvin Bridges, CNVs have been leveraged as molecular markers capable to guide animal breeding efforts. Recently, CNVs have been used to facilitate optimized cattle and sheep breeding. For example, MFN1 gene CNVs have been shown to influence Qinchuan cattle body size [6], while FecB CNVs are closely related to fertility traits in sheep [7]. In yaks, CADM2 CNVs are reportedly correlated with body weight [8], whereas HPGDS CNVs are significantly associated with both body weight and body length [9].
Members of the molecules interacting with the CasL (MICAL) gene family, including MICAL proteins and homologous MICAL-Like proteins, play important roles in diverse physiological processes [10]. MICAL can directly and specifically regulate actin dynamics, promoting semaphorin-induced signal conversion to promote F-actin instability-induced neuronal rejection [11]. MICAL-Like2 (MICALL2) encodes a protein lacking the N-terminal flavin monooxygenase domain present in MICAL proteins that is also referred to as the junctional Rab13-binding (JRAB) protein, given that it can serve as a Rab13 effector protein [12]. Through its ability to regulate claudin, E-cadherin, and occludin trafficking to the plasma membrane, MICALL2 serves as an essential regulator of adherens junction and tight junction assembly [13]. MICALL2 can additionally directly bind to F-actin in a manner that promotes its bundling and stabilization, while further regulating neurite outgrowth and epithelial cell scattering. This gene may thus play an important role in mammalian skeletal muscle development.
The monoacylglycerol O-acyltransferase 2 (MOGAT2) gene encodes an enzyme responsible for catalyzing diacylglycerol synthesis from 2-monoacylglycerol and fatty acyl-CoA. It plays an essential role in the digestion, absorption, and metabolic processing of fat. Mice lacking MOGAT2 expression are reportedly resistant to obesity and high-fat diet-associated metabolic disorders [14]. In Duroc pigs, MOGAT2 CNVs have also been reported to impact lipid metabolism [15].
MICALL2 CNVs have previously been reported to be correlated with the body weight, length, and height of young Nanyang cattle, although this correlative relationship was weaker in adult cattle [16]. MOGAT2 CNVs have also been annotated in a recent whole-gene resequencing study in yaks [17]. In the present study, a quantitative real-time PCR (qPCR) approach was employed to detect MICALL2 and MOGAT2 CNVs in Ashidan yaks and to examine the relationship between these CNVs and yak growth traits. In addition, the expression profiles of MICALL2 and MOGAT2 were compared across a range of Ashidan yak tissues.
2. Materials and Methods
2.1. Sample Collection and Growth Trait Measurement
Blood samples (4 mL) were collected from 315 female yaks from Datong Farm, Qinghai Province, China. All yaks were healthy and raised under identical conditions at an altitude of 3200 m. Four phenotypic traits were measured for each yak in this study (body weight, withers height, body length, chest girth) at 6, 12, 18, and 30 months of age. Measurements were made in accordance with the standard Gilbert method [18]. In addition, samples of heart, liver, spleen, lung, kidney, muscle, and adipose tissue were collected from three 18-month-old male yaks which were kept in the same conditions and at the same age, they were used to assess MICALL2 and MOGAT2 expression patterns in these different tissues.
2.2. Nucleic Acid Isolation
An Easy Pure Blood Genomic DNA Kit (TransGen Biotech, Beijing, China) was used to isolate genomic DNA from yak blood samples, after which a Nanodrop 2000 spectrophotometer (ThermoFisher Scientific Inc., Waltham, MA, USA) was used to assess DNA quality and concentration values. TRIZOL (Transgen Biotch, Beijing, China) was used to extract RNA from heart, liver, spleen, lung, kidney, muscle, and adipose tissue samples, after which 1.2% agarose gel electrophoresis and a spectrophotometer were used to assess RNA concentrations and quality. A PrimeScript™ Reagent Kit with gDNA Eraser (TaKaRa Bio Inc., Dalian, China) was used to prepare cDNA from 1 μg of total RNA. All DNA was stored at −20 °C.
2.3. Primer Design and Validation
The online National Center for Biotechnology Information (NCBI) primer-BLAST tool was used to design primers for analyses of MICALL2 and MOGAT2 CNVs and gene expression (Table 1). Optimal primer temperatures were determined through PCR and 1.5% agarose gel electrophoresis analyses. Each individual reaction included 10 μL of 2× Accurate Taq Master Mix (Accurate Biotechnology, Hunan, China), 0.4 μL of each primer, 0.1 μL of DNA, and 8.2 μL of sterile H2O. Thermocycler settings were as follows: 94 °C for 30 s; 30 cycles of 98 °C for 10 s, 55–60 °C for 30 s, and 72 °C for 1 min; 72 °C for 2 min.

Table 1.
Primer sequences.
2.4. MICALL2 and MOGAT2 CNV and Tissue Expression Analyses
For analyses of MICALL2 and MOGAT2 CNVs, basic transcription factor 3 (BTF3) served as an internal reference gene [19]. Genomic DNA was stained with SYBR® Green Pro Taq HS (Accurate Biology, Hunan, China), after which CNVs were measured for these two target genes via qPCR using a LightCycler® 96 Instrument (Roche, Basel, Switzerland) [20]. Each reaction included 10 μL of SYBR® Green Pro Taq HS, 0.4 μL of each primer, 1 μL of DNA, and 8.2 μL of sterile H2O. Thermocycler settings were as follows: 90 °C for 30 s; 45 cycles of 95 °C for 5 s and 60 °C for 30 s; 95 °C for 5 s; 65 °C for 60 s; hold at 95 °C. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is frequently used as a normalization control in tissue analyses [21], and it was thus used as a reference gene for analyses of MICALL2 and MOGAT2 gene expression in individual tissues. All qPCR reactions were performed using the same conditions used for CNV detection. Analyses were repeated in triplicate, and results are reported as means ± standard deviation (SD).
2.5. Association Analyses
MICALL2 and MOGAT2 CNVs were classified as gains, losses, or normal (non-CNVs) using the method, where [22]. Relationships between these CNV types and yak growth traits were then analyzed using analyses of variance (ANOVAs, SPSS Version19, IBM, New York, NY, USA). A value < 2.5 was classified as loss, while = 2.5 was classified as normal, and > 2.5 was classified as gain. The following linear model was used to explore the relationship between CNVs and growth traits: , where Yj is the observed growth trait value, μ is the overall mean value for that growth trait, CNVj is the effect of the three CNV types on this phenotype, and ej is the random residual error.
3. Results
3.1. Associations between Ashidan Yak CNVs and Growth Traits
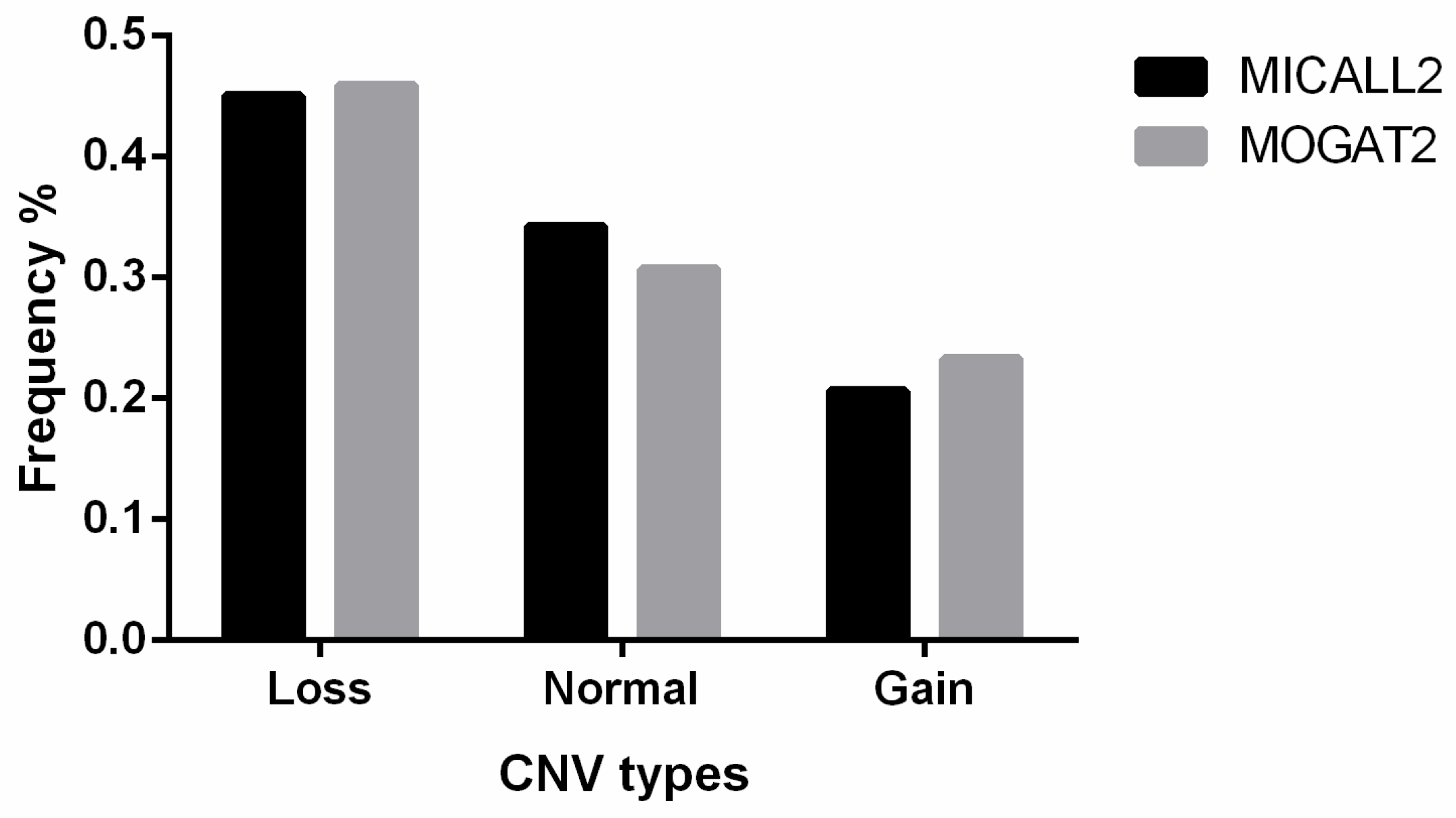
MICALL2 CNV classifications for the 315 sampled Ashidan yaks included 142, 108, and 65 loss-type, normal-type, and gain-type animals, respectively (Table 2). In addition, the MOGAT2 CNV classifications for 283 sampled Ashidan yaks included 130, 87, and 66 loss-type, normal-type, and gain-type animals, respectively (Figure 1).

Table 2.
Correlations between MICALL2 CNVs and Ashidan yak growth traits.
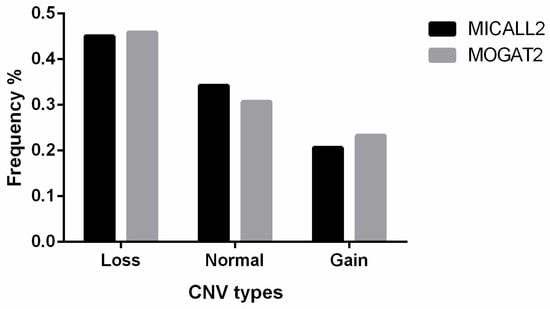
Figure 1.
MICALL2 and MOGAT2 gene CNV proportions in analyzed Ashidan yaks.
Correlation analyses revealed that MICALL2 and MOGAT2 CNVs were significantly correlated with certain growth traits in this yak population. Specifically, MICALL2 CNVs were significantly associated with body weight in 6-month-old yaks (p < 0.05), body length in 30-month-old yaks (p < 0.05), chest girth in 30-month-old yaks (p < 0.05), and withers height in 18-month-old yaks (p < 0.01) (Table 2). Similarly, MOGAT2 CNVs were significantly associated with body weight in 6-month-old and 30-month-old yaks (p < 0.05) and with withers height in 18-month-old yaks (p < 0.01) (Table 3)

Table 3.
Correlations between MOGAT2 CNVs and Ashidan yak growth traits.
3.2. MICALL2 and MOGAT2 Gene Expression Profiles
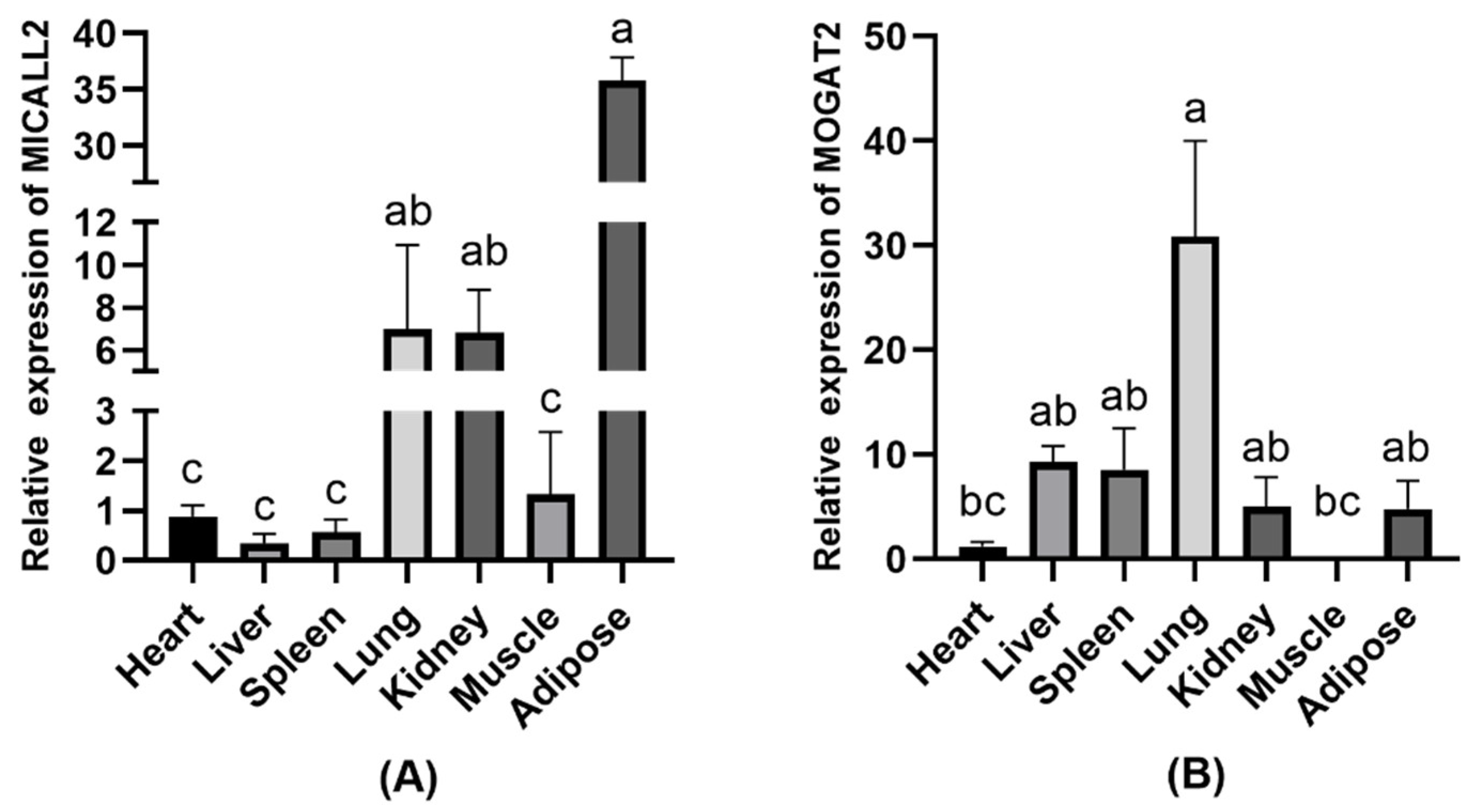
To validate the relationships between the expression of MICALL2 and MOGAT2 and the growth and development of Ashidan yaks, the mRNA expression profiles for these genes were analyzed in samples of Ashidan yak heart, liver, spleen, lung, kidney, muscle, and adipose tissue. These analyses revealed that MICALL2 was expressed at significantly higher levels in adipose tissue relative to lung or kidney tissue (p < 0.05) (Figure 2A). Moreover, MOGAT2 was expressed at significantly higher levels in lung samples relative to other tissues (p < 0.05) (Figure 2B).
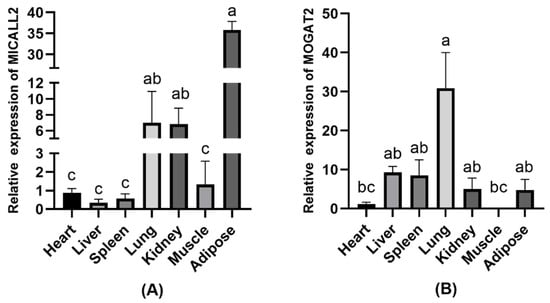
Figure 2.
MICALL2 and MOGAT2 expression in Ashidan yak tissue samples. (A) MICALL2 and (B) MOGAT2 expression levels in different tissues. a–c denote significant differences at p < 0.05.
4. Discussion
In recent years, there have been rapid advances in the field of molecular breeding, with marker-assisted selection (MAS) having accelerated the slower pace of more traditional breeding strategies [23]. In an analysis of 488 Chinese Qinchuan cattle, for example, researchers detected four single nucleotide polymorphisms (SNPs) in the CFL1 gene that were significantly correlated with key growth traits including body weight, body length, chest width, and chest depth [24]. Chickens exhibit high levels of NUDT15 expression in the bone tissue, and two insertion-deletion (InDel) variants in the promoter region of this gene have been linked to significant variability in chicken growth and carcass weight [25]. In sheep, TOP2B CNVs are also reportedly associated with the body length and chest circumference of sheep such that they may offer value for MAS-based breeding programs [26]. These genetic variations in livestock species can also influence breeding outcomes, with economically important traits such as meat quality and carcass weight being the primary focus of these analyses. CNVs in the SERPINA3-1 [27], DYNC1I2 [28], PLA2G2A [29], and SYT11 [30] genes can influence beef cattle growth traits, while CNVs in the CCSER1 [31], MYLK4 [32], PIGY [33], and KAT6A [34] genes can influence sheep and goat growth traits.
Here, Ashidan yaks served as experimental subjects, and CNVs in the MICALL2 and MOGAT2 genes were detected by qPCR to determine whether these variations were related to important yak growth traits. In addition, the expression profiles of these two genes were surveyed across a variety of yak tissues, ultimately supporting their potential effects on yak growth. The method [35] was used to normalize these data, with results being classified into three CNV categories (loss, gain, or normal). This approach ultimately revealed that MICALL2 CNVs were significantly correlated with body weight at 6 months of age, body length and chest girth at 30 months of age, and withers height at 18 months of age. Similarly, MOGAT2 CNVs were significantly associated with body weight at 6 and 30 months of age, and with withers height at 18 months of age in these Ashidan yaks. For the first three traits associated with MICALL2 CNVs, loss and normal classifications were associated with better phenotypic outcomes relative to gains in these yaks, whereas withers height values were higher for yaks exhibiting MICALL2 gains. We thus hypothesized that these MICALL2 CNVs adversely impact weight gain in young yaks, but positively impact the height of mature adult yaks while negatively impacting body length and chest girth. Similarly, MOGAT2 gains were significantly associated with poorer body weight in 6- and 30-month-old Ashidan yaks but were favorably associated with the withers height of 18-month-old yaks. In general, normal-type and gain-type were better than the loss-type, so the individuals with MICALL2 gene loss-type could be eliminated in yak breeding, and the growth traits of MOGAT2 gain-type were also generally worse than those of loss-type and normal-type, these guesses need to be confirmed by subsequent experiments. The two gene results require further experimental validation. Gene expression profiling confirmed that MICALL2 was expressed at high levels in adipose, lung, and kidney tissues, while MOGAT2 was highly expressed in lung tissue samples. As such, these genes may govern the growth and development of these target tissues in Ashidan yaks.
Although initially identified in mammals, the MICAL protein family has primarily been characterized in studies of Drosophila model animals [11]. These MICAL family members can reportedly interact with F-actin and influence cytoskeletal development. MICAL gene expression has been detected in both embryonic and adult nervous system tissues [36]. High levels of MICALL2 expression have been reported in a range of malignancies including ovarian, gastric, and breast cancers, consistent with the high levels of variability observed for this gene [37]. This suggests that this gene can readily undergo mutation and that it can affect adipose and nervous system tissue development. As such, MICALL2 CNVs in yaks may similarly influence adipose tissue development in a manner that ultimately alters the growth traits observed in these yaks.
MOGAT2 encodes an enzyme responsible for triacylglycerol resynthesis, which is critical to fat absorption in the small intestine. Given its role in the absorption of dietary fat, MOGAT2 has been established as an important driver of obesity in humans [38]. MOGAT2 deficiency results in increased energy expenditure and the suppression of weight gain in a genetic mouse model of obesity [39]. In yaks, however, fat intake and storage are both beneficial adaptations to the harsh plateau environment. Two studies have previously identified MOGAT2 CNVs in yaks and Duroc pics through whole-genome sequencing and annotated the function of this gene [15,17]. Accordingly, the association between MOGAT2 CNVs and yak growth traits was herein analyzed. In Ashidan yaks, these MOGAT2 CNVs were associated with several growth traits including body weight and body height, potentially mediating these effects through increased fat absorption and consequent increases in yak growth performance. These CNVs may also influence lung development, thereby impacting blood oxygen content or other factors that have the potential to impact overall yak growth and development [40].
5. Conclusions
In summary, these results offer new evidence that CNVs in the MICALL2 and MOGAT2 genes are associated with yak growth traits. As such, these findings provide a robust theoretical foundation for the use of these two genes as targets for molecular breeding efforts aimed at improving Ashidan yak growth performance.
Author Contributions
Conceptualization, M.L., C.L. and P.Y.; methodology, M.L. and X.W.; software, M.L. and R.D.; validation, M.L., C.H. and W.R.; formal analysis, M.L. and X.M.; investigation, M.L. and X.W.; resources, M.C. and P.B.; data curation, M.L.; writing—original draft preparation, M.L. and L.X.; writing—review and editing, M.L. and X.L.; visualization, M.L. and J.P.; supervision, X.G.; project administration, C.L.; funding acquisition, P.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by State Key R & D program (2021YFD1600200); Gansu basic research innovation group project (20JR5RA580); Major science and technology projects in Gansu Province (21ZD10NA001, GZGG-2021-1); Modern beef yak industry technology system (MATS-Beef Cattle System, CARS-37); Yak resources and breeding innovation project of Chinese Academy of Agricultural Sciences (25-LIHPS-01).
Institutional Review Board Statement
All experiments in this study were approved by the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS, the grant number is NO. LIHPS-CAAS-2017-115. Additionally, we collected all of the blood samples and analyzed the data strictly following the guidelines for the Care and Use of Laboratory Animals by the People’s Republic of China.
Informed Consent Statement
The Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS approved all animal studies (approval NO. LIHPS-CAAS-2017-115). All blood and tissue samples were collected and data analyses were performed in strict accordance with the guidelines for the Care and Use of Laboratory Animals established by the People’s Republic of China, samples were also taken with the consent of the farmer and related personnel.
Data Availability Statement
The study did not report any data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prakash, B.S.; Sarkar, M.; Mondal, M. An update on reproduction in yak and mithun. Reprod. Domest. Anim. Zuchthyg. 2008, 43 (Suppl. S2), 217–223. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, X.; Ding, X.; Chu, M.; Liang, C.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. The Selection of Reference Genes for Quantitative Real-Time PCR in the Ashidan Yak Mammary Gland During Lactation and Dry Period. Animals 2019, 9, 943. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hu, Q.; Ma, H.; Wang, L.; Yang, Y.; Luo, W.; Qiu, Q. Genome-wide variation within and between wild and domestic yak. Mol. Ecol. Resour. 2014, 14, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Macé, A.; Kutalik, Z.; Valsesia, A. Copy Number Variation. Methods Mol. Biol. 2018, 1793, 231–258. [Google Scholar]
- Upadhyay, M.; da Silva, V.H.; Megens, H.J.; Visker, M.; Ajmone-Marsan, P.; Bâlteanu, V.A.; Dunner, S.; Garcia, J.F.; Ginja, C.; Kantanen, J.; et al. Distribution and Functionality of Copy Number Variation across European Cattle Populations. Front. Genet. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Li, J.; Zhang, Z.; Chai, Y.; Liu, X.; Li, J.; Huang, Y.; Li, L.; Huang, W.; Yang, G.; et al. The relationship between MFN1 copy number variation and growth traits of beef cattle. Gene 2022, 811, 146071. [Google Scholar] [CrossRef]
- Bi, Y.; Feng, W.; Kang, Y.; Wang, K.; Yang, Y.; Qu, L.; Chen, H.; Lan, X.; Pan, C. Detection of mRNA Expression and Copy Number Variations Within the Goat Fec (B) Gene Associated With Litter Size. Front. Vet. Sci. 2021, 8, 758705. [Google Scholar] [CrossRef]
- Ge, F.; Jia, C.; Chu, M.; Liang, C.; Yan, P. Copy Number Variation of the CADM2 Gene and Its Association with Growth Traits in Yak. Animal 2019, 9, 1008. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Ren, W.; Zhang, Y.; Wu, X.; Zhang, Q.; Ma, X.; Bao, P.; Guo, X.; Chu, M.; et al. Copy number variation of the HPGDS gene in the Ashidan yak and its associations with growth traits. Gene 2021, 772, 145382. [Google Scholar] [CrossRef]
- Min, P.; Zhang, L.; Wang, Y.; Qi, C.; Song, Y.; Bibi, M.; Zhang, Y.; Ma, Y.; Zhao, X.; Yu, M.; et al. MICAL-L2 Is Essential for c-Myc Deubiquitination and Stability in Non-small Cell Lung Cancer Cells. Front. Cell Dev. Biol. 2020, 8, 575903. [Google Scholar] [CrossRef]
- Giridharan, S.S.; Rohn, J.L.; Naslavsky, N.; Caplan, S. Differential regulation of actin microfilaments by human MICAL proteins. J. Cell Sci. 2012, 125, 614–624. [Google Scholar] [CrossRef]
- Rahajeng, J.; Giridharan, S.S.; Cai, B.; Naslavsky, N.; Caplan, S. Important relationships between Rab and MICAL proteins in endocytic trafficking. World J. Biol. Chem. 2010, 1, 254–264. [Google Scholar] [CrossRef]
- Giridharan, S.S.; Caplan, S. MICAL-family proteins: Complex regulators of the actin cytoskeleton. Antioxid. Redox Signal. 2014, 20, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Nelson, D.W.; Banh, T.; Yen, M.I.; Yen, C.E. Intestine-specific expression of MOGAT2 partially restores metabolic efficiency in Mogat2-deficient mice. J. Lipid Res. 2013, 54, 1644–1652. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, Y.; Liu, S.; Meng, Q. Genome-Wide Assessment Characteristics of Genes Overlapping Copy Number Variation Regions in Duroc Purebred Population. Front. Genet. 2021, 12, 753748. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.; Shi, T.; Zhou, Y.; Cai, H.; Lan, X.; Zhang, C.; Lei, C.; Chen, H. Copy number variations of MICAL-L2 shaping gene expression contribute to different phenotypes of cattle. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2013, 24, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chai, Z.; Hu, D.; Ji, Q.; Xin, J.; Zhang, C.; Zhong, J. A global analysis of CNVs in diverse yak populations using whole-genome resequencing. BMC Genom. 2019, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.P.; Bailey, D.R.; Shannon, N.H. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. J. Anim. Sci. 1993, 71, 1712–1720. [Google Scholar] [CrossRef]
- Ali, S.; Srivastava, A.K.; Chopra, R.; Aggarwal, S.; Garg, V.K.; Bhattacharya, S.N.; Bamezai, R.N. IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J. Med. Genet. 2013, 50, 34–42. [Google Scholar] [CrossRef]
- Xie, C.; Tammi, M.T. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinform. 2009, 10, 80. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Dube, S.; Mir, A.; Qin, J.; Sun, G.; Ramakrishnan, R.; Jones, R.C.; Livak, K.J. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 2010, 50, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.L. The use of marker-assisted selection in animal breeding and biotechnology. Rev. Sci. Et Tech. Int. Off. Epizoot. 2005, 24, 379–391. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, C.; Yang, Z.; Li, M.; Chen, Z.; Xu, T.; Zhang, H.; Mao, Y. The polymorphism of bovine Cofilin-1 gene sequence variants and association analysis with growth traits in Qinchuan cattle. Anim. Biotechnol. 2022, 33, 63–69. [Google Scholar] [CrossRef]
- Wei, C.; Niu, Y.; Chen, B.; Qin, P.; Wang, Y.; Hou, D.; Li, T.; Li, R.; Wang, C.; Yin, H.; et al. Genetic effect of an InDel in the promoter region of the NUDT15 and its effect on myoblast proliferation in chickens. BMC Genom. 2022, 23, 138. [Google Scholar] [CrossRef] [PubMed]
- Toremurat, Z.; Ibrahim, E.E.; Huang, Y.Z.; Lan, X.; Pi, L.; Chaogetu, B.; Hu, L.; Chen, H. Copy number variations of TOP2B gene are associated with growth traits in Chinese sheep breeds. Anim. Biotechnol. 2022, 33, 85–89. [Google Scholar] [CrossRef]
- Huang, Y.Z.; Shi, Q.T.; Shi, S.Y.; Yang, P.; Zhang, Z.J.; Lyu, S.J.; Chen, F.Y.; Xu, J.W.; Liu, X.; Li, Z.; et al. Association between copy number variation of SERPINA3-1 gene and growth traits in Chinese cattle. Anim. Biotechnol. 2022, 24, 1–8. [Google Scholar] [CrossRef]
- Li, X.; Ding, X.; Liu, L.; Yang, P.; Yao, Z.; Lei, C.; Chen, H.; Huang, Y.; Liu, W. Copy number variation of bovine DYNC1I2 gene is associated with body conformation traits in chinese beef cattle. Gene 2022, 810, 146060. [Google Scholar] [CrossRef]
- Yang, P.; Cai, C.; Niu, M.; Liu, X.; Wang, H.; Liang, H.; Cheng, B.; Zhang, Z.; Chen, F.; Xie, J.; et al. Effect of copy number variation of PLA2G2A gene to growth traits in Chinese cattle. Gene 2022, 809, 146014. [Google Scholar] [CrossRef]
- Yang, H.; Yue, B.; Yang, Y.; Tang, J.; Yang, S.; Qi, A.; Qu, K.; Lan, X.; Lei, C.; Wei, Z.; et al. Distribution of Copy Number Variation in SYT11 Gene and Its Association with Growth Conformation Traits in Chinese Cattle. Biology 2022, 11, 223. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, X.; Song, X.; An, Q.; Wang, D.; Zhang, Z.; Ding, X.; Yao, Z.; Wang, E.; Liu, X.; et al. Association between the copy number variation of CCSER1 gene and growth traits in Chinese Capra hircus (goat) populations. Anim. Biotechnol. 2022, 2, 1–7. [Google Scholar] [CrossRef]
- Shi, S.Y.; Li, L.J.; Zhang, Z.J.; Wang, E.Y.; Wang, J.; Xu, J.W.; Liu, H.B.; Wen, Y.F.; He, H.; Lei, C.Z.; et al. Copy number variation of MYLK4 gene and its growth traits of Capra hircus (goat). Anim. Biotechnol. 2020, 31, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, X.; Cheng, J.; Jiang, R.; Huang, R.; Wang, D.; Huang, Y.; Pi, L.; Hu, L.; Chen, H. Copy Number Variation of the PIGY Gene in Sheep and Its Association Analysis with Growth Traits. Animal 2020, 10, 688. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Cao, X.; Huang, Y.; Li, P.; Lan, X.; Buren, C.; Hu, L.; Chen, H. Copy number variations of the KAT6A gene are associated with body measurements of Chinese sheep breeds. Anim. Biotechnol. 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pasterkamp, R.J.; Dai, H.N.; Terman, J.R.; Wahlin, K.J.; Kim, B.; Bregman, B.S.; Popovich, P.G.; Kolodkin, A.L. MICAL flavoprotein monooxygenases: Expression during neural development and following spinal cord injuries in the rat. Mol. Cell. Neurosci. 2006, 31, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, M.S.; Bell, E.S.; Girard, M.; Chaineau, M.; Hamlin, J.N.; Daubaras, M.; Monast, A.; Park, M.; Hodgson, L.; McPherson, P.S. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J. Cell Biol. 2015, 208, 629–648. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.L.; Farese, R.V., Jr. MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine. J. Biol. Chem. 2003, 278, 18532–18537. [Google Scholar] [CrossRef]
- Nelson, D.W.; Gao, Y.; Spencer, N.M.; Banh, T.; Yen, C.L. Deficiency of MGAT2 increases energy expenditure without high-fat feeding and protects genetically obese mice from excessive weight gain. J. Lipid Res. 2011, 52, 1723–1732. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Wang, X.; Jiang, Q.; Zhao, H.; Wang, J.; Ju, Z.; Yang, L.; Gao, Y.; Wei, X.; et al. Population Structure, and Selection Signatures Underlying High-Altitude Adaptation Inferred From Genome-Wide Copy Number Variations in Chinese Indigenous Cattle. Front. Genet. 2019, 10, 1404. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).