Ikhnos: A Novel Software to Register and Analyze Bone Surface Modifications Based on Three-Dimensional Documentation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ikhnos and IkhnosToolBox
2.2. Sample
2.3. Statistical Analyses
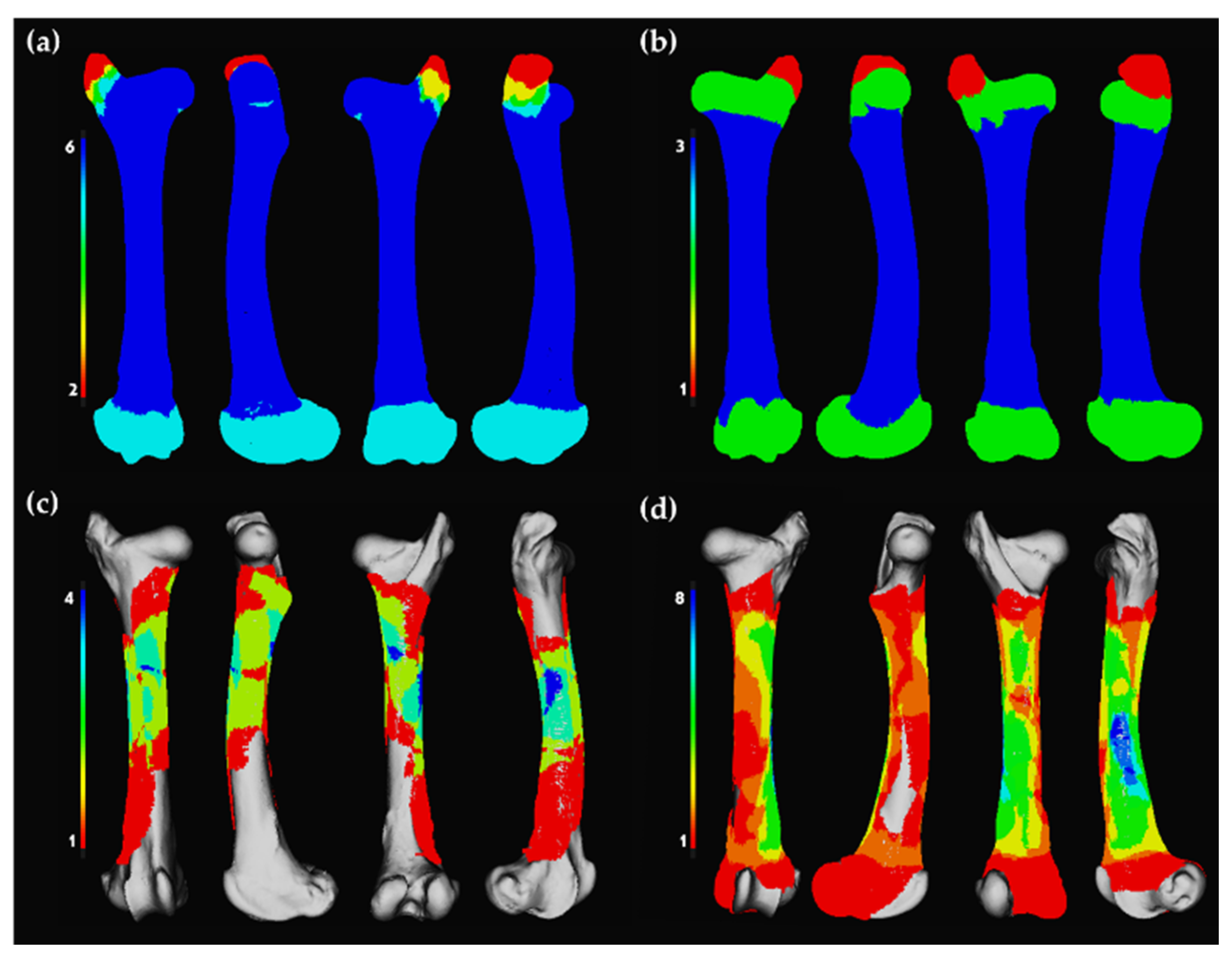
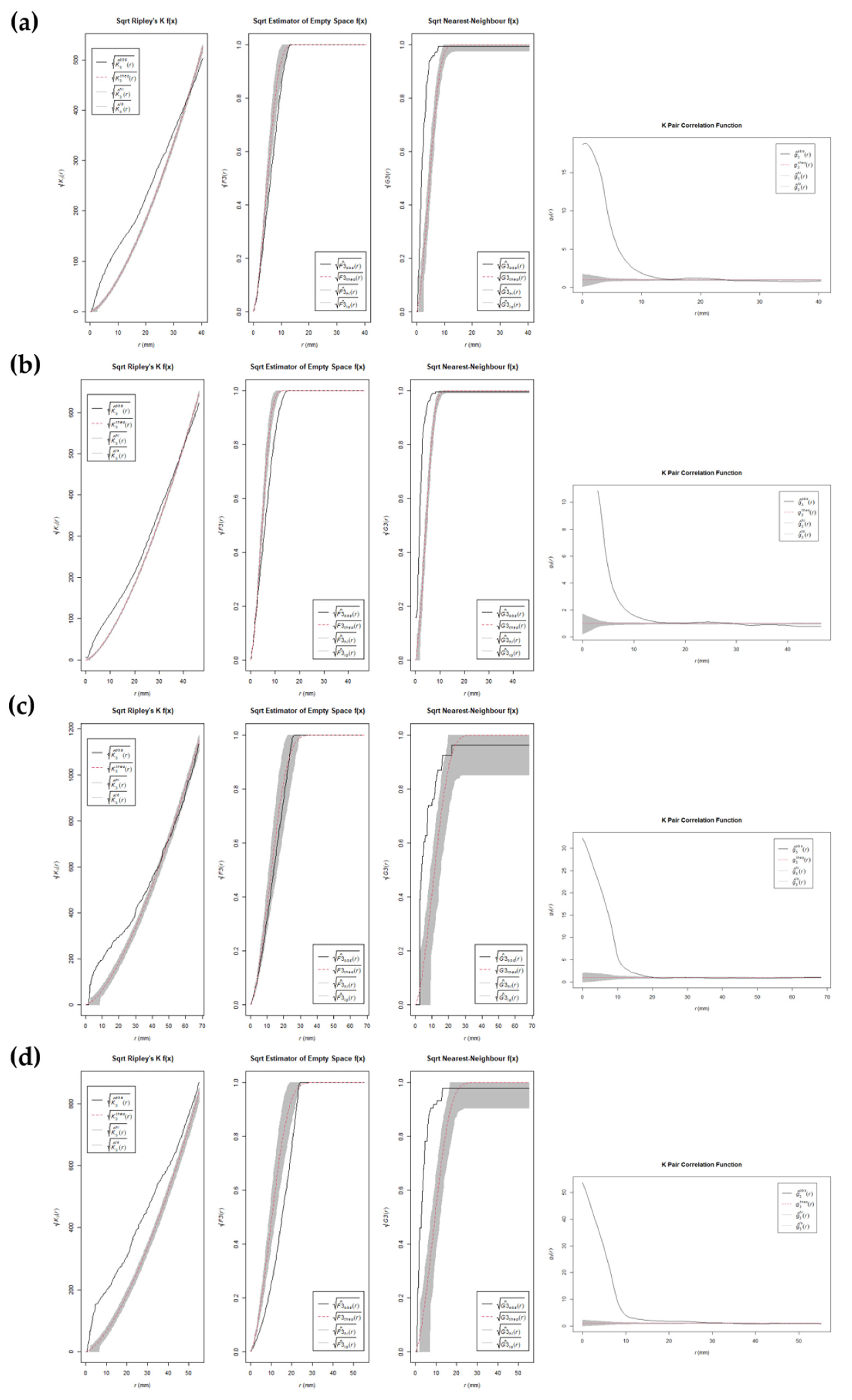
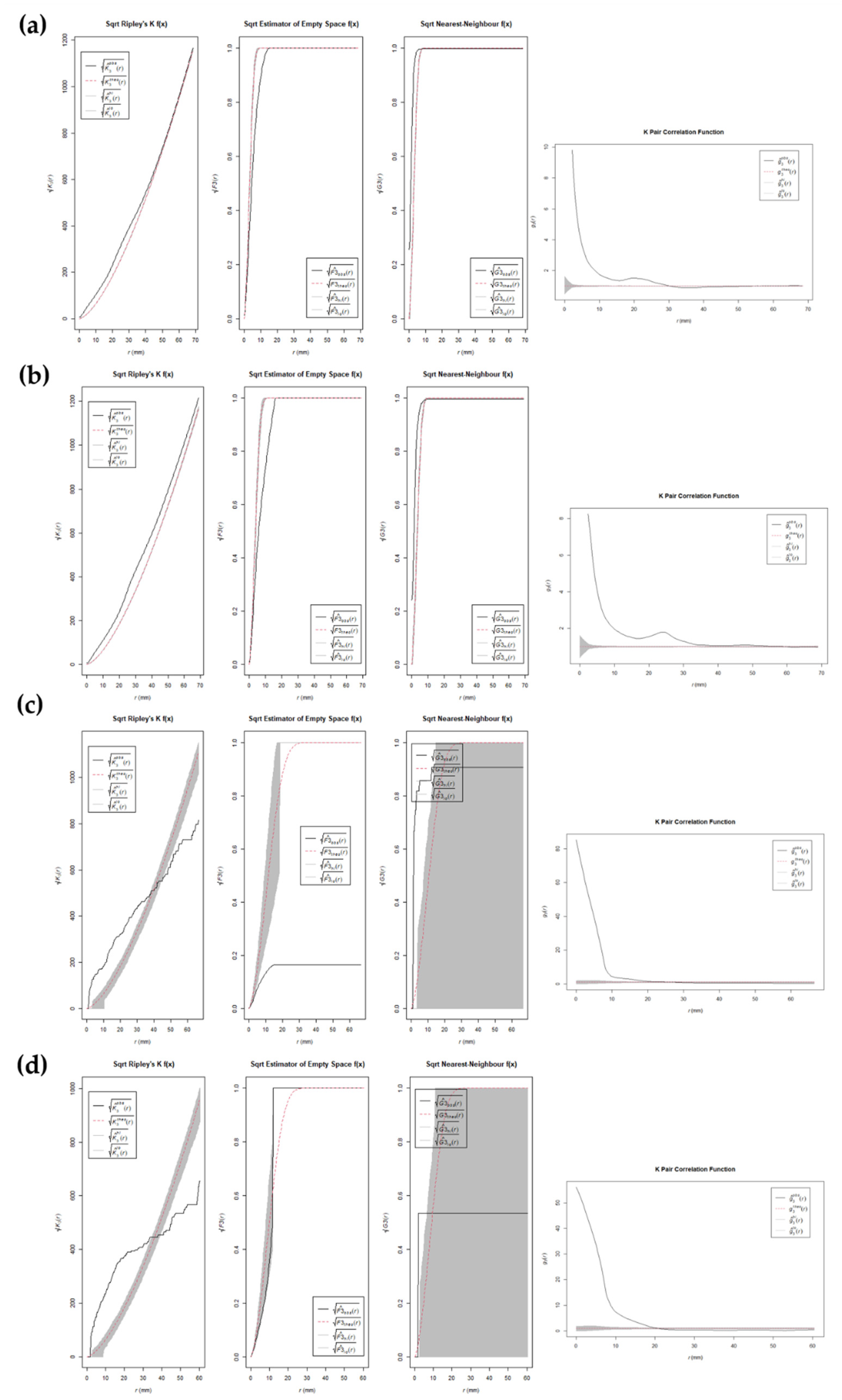
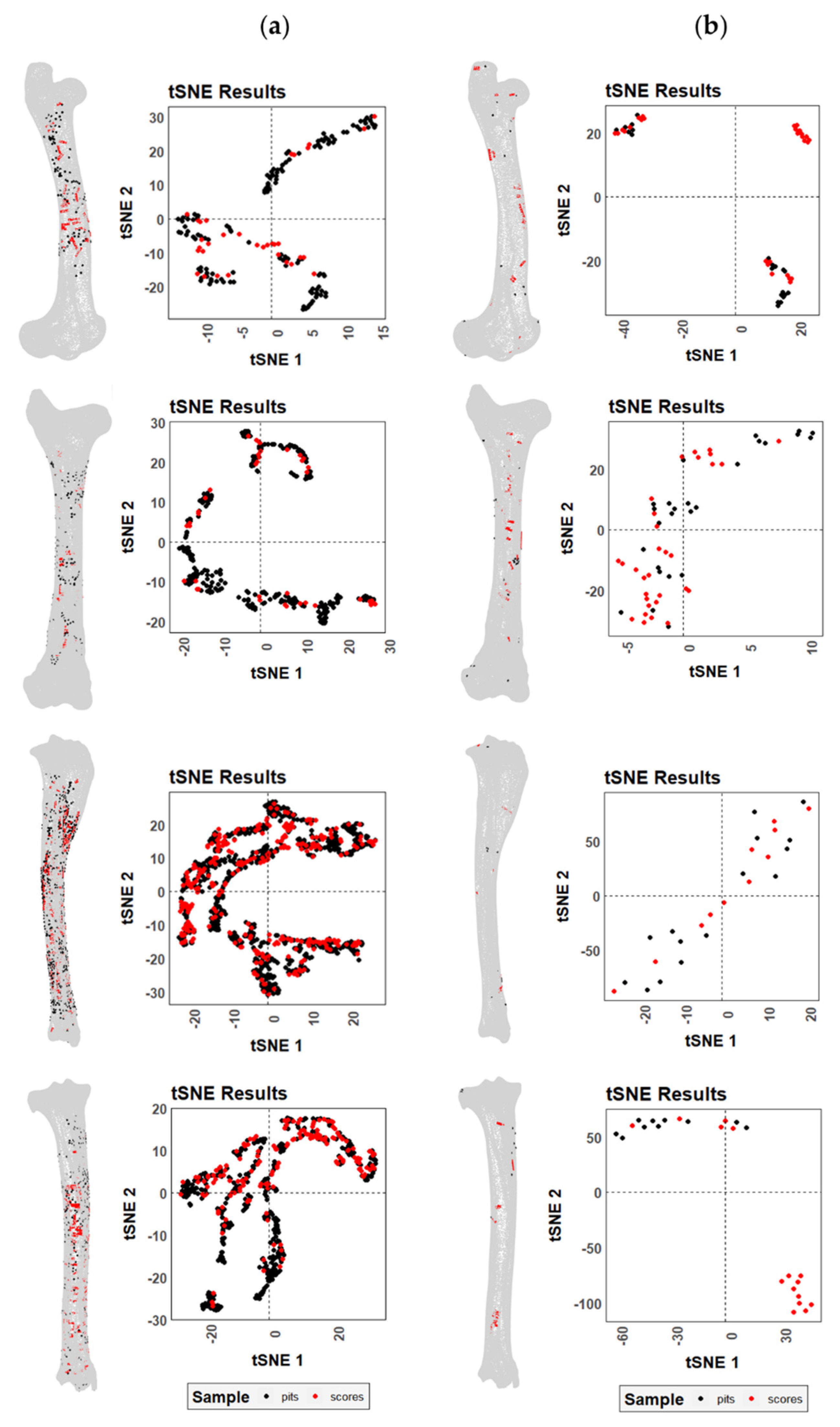
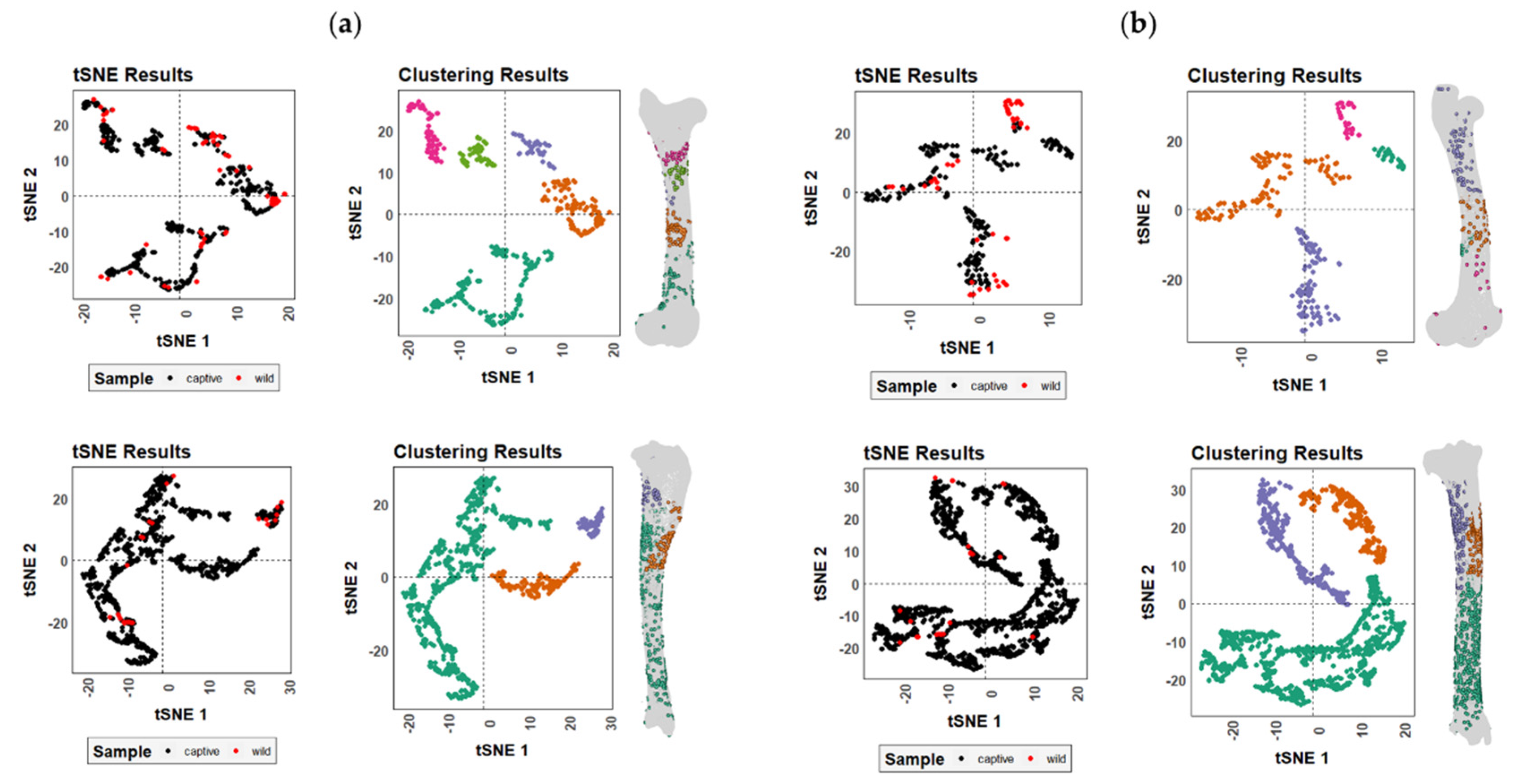
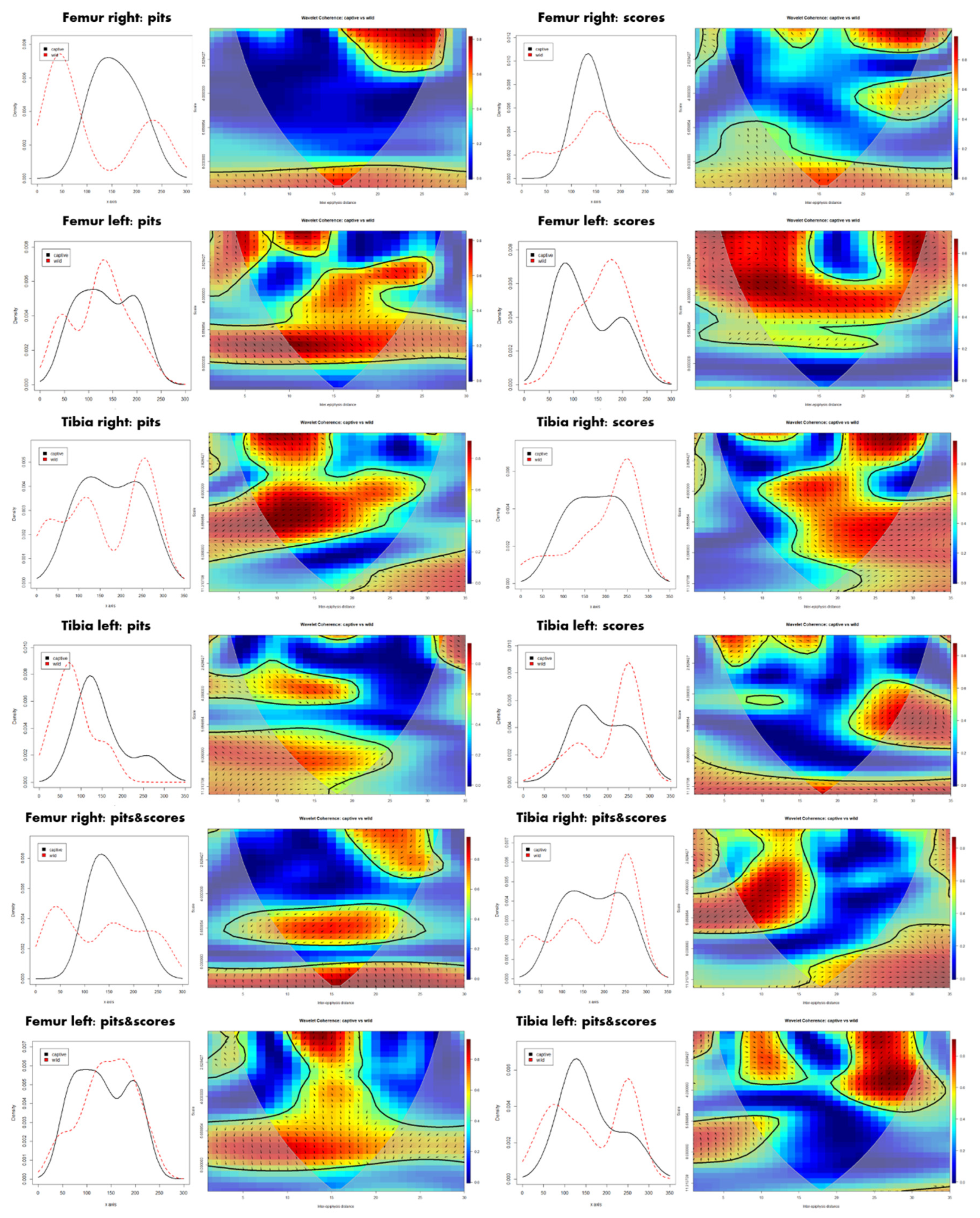
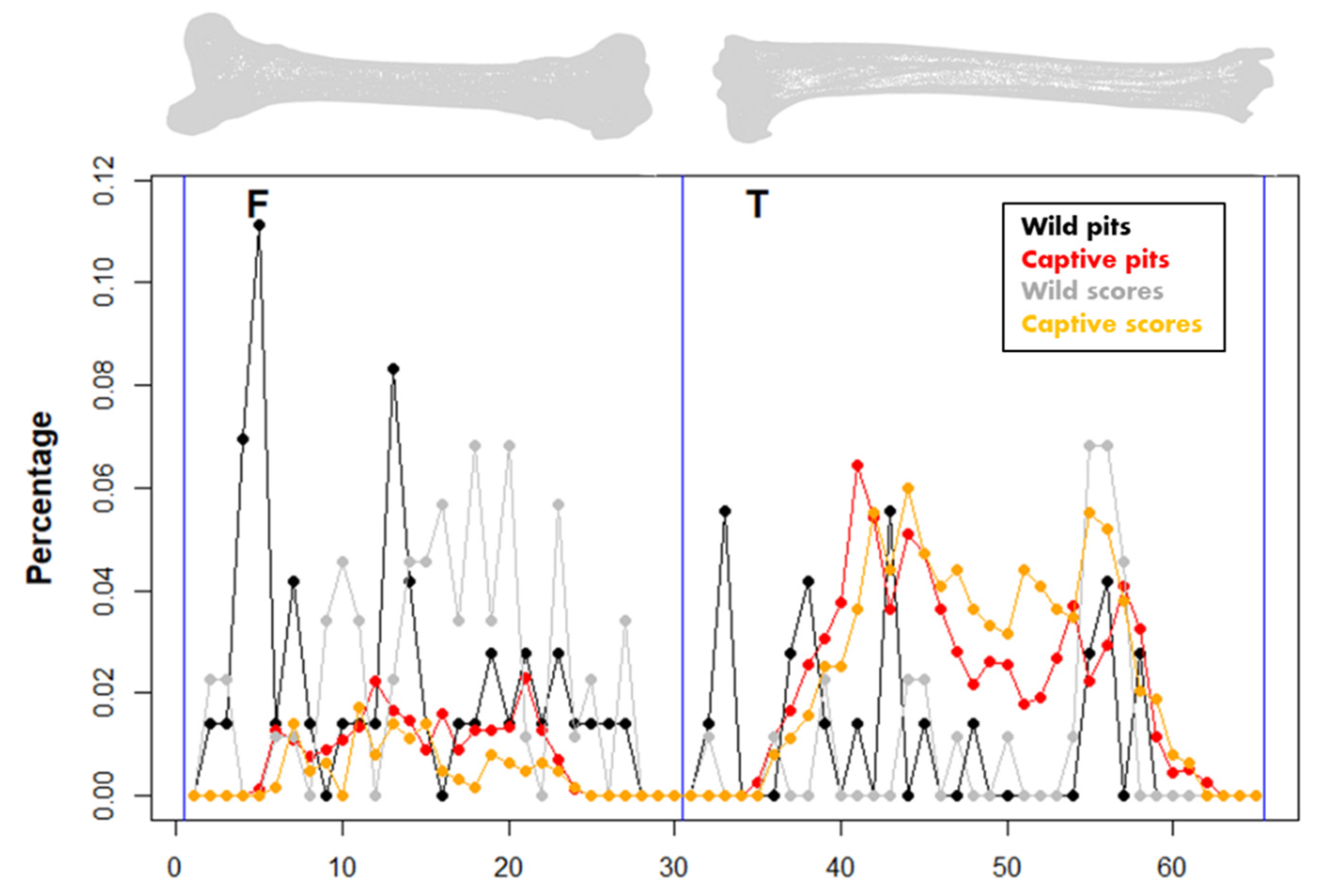
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blumenschine, R.J.; Marean, C.W.; Capaldo, S.D. Blind tests of inter-analyst correspondence and accuracy in the identification of cut marks, percussion marks, and carnivore tooth marks on bone surfaces. J. Archaeol. Sci. 1996, 23, 493–507. [Google Scholar] [CrossRef]
- Blumenschine, R.J.; Selvaggio, M.M. Percussion marks on bone surfaces as a new diagnostic of hominid behaviour. Nature 1988, 333, 763–765. [Google Scholar] [CrossRef]
- Selvaggio, M.M.; Wilder, J. Identifying the Involvement of Multiple Carnivore Taxa with Archaeological Bone Assemblages. J. Archaeol. Sci. 2001, 28, 465–470. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; de Juana, S.; Galán, A.B.; Rodríguez, M. A new protocol to differentiate trampling marks from butchery cut marks. J. Archaeol. Sci. 2009, 36, 2643–2654. [Google Scholar] [CrossRef]
- Madgwick, R. What makes bones shiny? Investigating trampling as a cause of bone abrasion. Archaeol. Anthropol. Sci. 2014, 6, 163–173. [Google Scholar] [CrossRef]
- Njau, J.K.; Blumenschine, R.J. A diagnosis of crocodile feeding traces on larger mammal bone, with fossil examples from the Plio-Pleistocene Olduvai Basin, Tanzania. J. Hum. Evol. 2006, 50, 142–162. [Google Scholar] [CrossRef]
- Oliver, J.S. Estimates of hominid and carnivore involvement in the FLK Zinjanthropus fossil assemblage: Some socioecological implications. J. Hum. Evol. 1994, 27, 267–294. [Google Scholar] [CrossRef]
- Pickering, T.R.; Dominguez-Rodrigo, M.; Egeland, C.P.; Brain, C.K. Beyond leopards: Tooth marks and the contribution of multiple carnivore taxa to the accumulation of the Swartkans Member 3 fossil assemblage. J. Hum. Evol. 2004, 46, 595–604. [Google Scholar] [CrossRef]
- Pobiner, B.L. Hominin–Carnivore Interactions: Evidence from Modern Carnivore Bone Modification and Early Pleistocene Archaeofaunas (Koobi Fora, Kenya; Olduvai Gorge, Tanzania). Ph.D. Dissertation, Rutgers University, New Brunswick, NJ, USA, 2007. [Google Scholar]
- Shipman, P.; Rose, J. Early hominid hunting, butchering, and carcass-processing behaviours: Approaches to the fossil record. J. Anthropol. Archaeol. 1983, 2, 57–98. [Google Scholar] [CrossRef]
- Shipman, P.; Fisher, D.; Rose, J. Mastodon butchery: Microscopic evidence of carcass processing and bone tool use. Paleobiology 1984, 10, 358–365. [Google Scholar] [CrossRef]
- Bello, S.M.; de Groote, I.; Delbarre, G. Application of 3-dimensional microscopy and micro-CT scanning to the analysis of Magdalenian portable art on bone and antler. J. Archaeol. Sci. 2013, 40, 2464–2476. [Google Scholar] [CrossRef]
- Bello, S.M.; Galway-Witham, J. Bone taphonomy inside and out: Application of 3-dimensional microscopy, scanning electron microscopy and micro-computed tomography to the study of humanly modified faunal assemblages. Quat. Int. 2019, 517, 16–32. [Google Scholar] [CrossRef]
- Delaney-Rivera, C.; Plummer, T.W.; Hodgson, J.A.; Forrest, F.; Hertel, F.; Oliver, J.S. Pits and pitfalls: Taxonomic variability and patterning in tooth mark dimensions. J. Archaeol. Sci. 2009, 36, 2597–2608. [Google Scholar] [CrossRef]
- Landt, M.J. Tooth Marks and human consumption: Ethnoarchaeological mastication research among foragers of the Central African Republic. J. Archaelogical Sci. 2007, 34, 1629–1640. [Google Scholar] [CrossRef]
- Maté-González, M.A.; Yravedra, J.; González-Aguilera, D.; Palomeque-González, J.F.; Domínguez-Rodrigo, M. Micro-photogrammetric characterization of cut marks on bones. J. Archaeol. Sci. 2015, 62, 128–142. [Google Scholar] [CrossRef]
- Maté-González, M.A.; Aramendi, J.; Yravedra, J.; Blasco, R.; Rosell, J.; González-Aguilera, D.; Domínguez-Rodrigo, M. Assessment of statistical agreement of three techniques for the study of cut marks: 3D digital microscope, laser scanning confocal microscopy and micro-photogrammetry. J. Microsc. 2017, 267, 356–370. [Google Scholar] [CrossRef]
- Aramendi, J.; Maté-González, M.A.; Yravedra, J.; Ortega, M.C.; Arriaza, M.C.; González-Aguilera, D.; Baquedano, E.; Domínguez-Rodrigo, M. Discerning carnivore agency through the three-dimensional study of tooth pits: Revisiting crocodile feeding behaviour at FLK-Zinj and FLK NN3 (Olduvai Gorge, Tanzania). Palaeogeogr. Paleoclimatol. Palaeoecol. 2017, 488, 93–102. [Google Scholar] [CrossRef]
- Arriaza, M.C.; Aramendi, J.; Maté-González, M.A.; Yravedra, J.; Stratford, D. Characterising leopard as taphonomic agent through the use of micro-photogrammetric reconstruction of tooth marks and pit to score ratio. Hist. Biol. 2019, 33, 176–185. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Maté-González, M.A.; Aramendi, J.; González-Aguilera, D. 3D analysis of cut marks using a new geometric morphometric methodological approach. Archaeol. Anthropol. Sci. 2019, 11, 651–665. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Huguet, R.; Yravedra, J. Scratches and grazes: A detailed microscopic analysis of trampling phenomena. J. Microsc. 2020, 277, 107–117. [Google Scholar] [CrossRef]
- Otárola-Castillo, E.; Torquato, M.G.; Hawkins, H.C.; James, E.; Harris, J.A.; Marean, C.W.; McPherron, S.P.; Thomson, J.C. Differentiating between cutting actions on bone using 3D geometric morphometrics and Bayesian analyses with implications to human evolution. J. Archaeol. Sci. 2018, 89, 56–67. [Google Scholar] [CrossRef]
- Souron, A.; Napias, A.; Lavidalie, T.; Santos, F.; Ledevin, R.; Castel, J.-C.; Costamagno, S.; Cusimano, D.; Drumheller, S.; Parkinson, J.M.; et al. A new geometric morphometrics-based shape and size analysis discriminating anthropogenic and non-anthropogenic bone surface modifications of an experimental data set. In Proceedings of the IMEKO TC-4 International Conference on Metrology for Archaeology and Cultural Heritage, Florence, Italy, 4–6 December 2019. [Google Scholar]
- Yravedra, J.; Aramendi, J.; Maté-González, M.A.; Courtenay, L.A.; González-Aguilera, D. Differentiating percussion pits and carnivore tooth pits using 3D reconstructions and geometric morphometrics. PLoS ONE 2018, 13, e0194324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cifuentes-Alcobendas, G.; Domínguez-Rodrigo, M. Deep learning and taphonomy: High accuracy in the classification of cut marks made on fleshed and defleshed bones using convolutional neural networks. Sci. Rep. 2019, 9, 18933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cifuentes-Alcobendas, G.; Domínguez-Rodrigo, M. More than meets the eye: Use of computer vision algorithms to identify stone tool material through the analysis of cut mark micro-morphology. Archaeol. Anthropol. Sci. 2021, 13, 167. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Huguet, R.; Aramendi, J.; Maté-González, M.A.; González-Aguilera, D.; Arriaza, M.C. Combining machine learning algorithms and geometric morphometrics: A study of carnivore tooth marks. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2019, 522, 28–39. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Huguet, R.; González-Aguilera, D.; Yravedra, J. A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification. Appl. Sci. 2019, 10, 150. [Google Scholar] [CrossRef] [Green Version]
- Courtenay, L.A.; Herranz-Rodrigo, D.; González-Aguilera, D.; Yravedra, J. Developments in data science solutions for carnivore tooth pit classification. Sci. Rep. 2021, 11, 10209. [Google Scholar] [CrossRef]
- Blumenschine, R.J. Percussion marks, tooth marks, and experimental determinations of the timing of hominid and carnivore access to long bones at FLK Zinjanthropus, Olduvai Gorge, Tanzania. J. Hum. Evol. 1995, 29, 21–51. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Bunn, H.T.; Yravedra, J. A critical re-evaluation of bone surface modification models for inferring fossil hominin and carnivore interactions through a multivariate approach: Application to the FLK Zinj archaeofaunal assemblage (Olduvai Gorge, Tanzania). Quat. Int. 2014, 322–323, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Pante, M.C.; Blumenschine, R.J.; Capaldo, S.D.; Scott, R.S. Validation of bone surface modification models for inferring fossil hominin and carnivore feeding interactions, with reapplication to FLK 22, Olduvai Gorge, Tanzania. J. Hum. Evol. 2012, 63, 395–407. [Google Scholar] [CrossRef]
- Selvaggio, M.M. Carnivore tooth marks and stone tool butchery marks on scavenged bones: Archaeological implications. J. Hum. Evol. 1994, 27, 215–228. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M. Flesh availability and bone modification in carcasses consumed by lions. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1999, 149, 373–388. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M. Testing meat-eating in early hominids: An analysis of butchery marks on defleshed carcases. Hum. Evol. 1997, 12, 169–182. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M. Meat-eating by early hominids at the FLK 22 Zinjanthropus site, Olduvai Gorge (Tanzania): An experimental approach using cut-mark data. J. Hum. Evol. 1997, 33, 669–690. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Rodrigo, M.; Pickering, T.R. Early Hominid Hunting and Scavenging: A Zooarchaeological Review. Evol. Anthropol. 2003, 12, 275–282. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Barba, R.; Egeland, C.P. Deconstructing Olduvai: A Taphonomic Study of the Bed I Sites, 1st ed.; Springer: Dordrecht, The Netherlands, 2007; p. 337. [Google Scholar]
- Domínguez-Rodrigo, M.; Barba, R. New estimates of tooth mark and percussion mark frequencies at the FLK Zinj site: The carnivore-hominid-carnivore hypothesis falsified. J. Hum. Evol. 2006, 50, 170–194. [Google Scholar] [CrossRef]
- Galán, A.B.; Domínguez-Rodrigo, M. An experimental study of the anatomical distribution of cut marks created by filleting and disarticulation of long bone ends. Archaeometry 2013, 55, 1132–1149. [Google Scholar] [CrossRef]
- Abe, Y.; Curtis, M.W.; Nilssen, P.J.; Assefa, Z.; Stone, E.C. The Analysis of Cutmarks on Archaeofauna: A Review and Critique of Quantification Procedures, and a New Image-Analysis GIS Approach. Am. Antiq. 2002, 67, 643–663. [Google Scholar] [CrossRef]
- Ciesielski, E.; Bohbot, H. Analyses of Bone Modifications on Human Remains: A GIS Approach. In CAA2014. 21st Century Archaeology. Concepts, Methods and Tools. Proceeding of the 42nd Annual Conference on Computer Applications and Quantitative Methods in Archaeology, Paris, France, 22–25 April 2014. [Google Scholar]
- Marginedas, F.; Rodríguez-Hidalgo, A.; Soto, M.; Bello, S.M.; Cáceres, I.; Huguet, R.; Saladié, P. Making skull cups: Butchering traces on cannibalised human skulls from five European archaeological sites. J. Archaeol. Sci. 2020, 114, 105076. [Google Scholar] [CrossRef]
- Parkinson, J.A. Revisiting the hunting-versus-scavenging debate at FLK-Zinj: A GIS spatial analysis of bone surface modifications produced by hominins and carnivores in the FLK 22 assemblage, Olduvai Gorge, Tanzania. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2018, 511, 29–51. [Google Scholar] [CrossRef]
- Parkinson, J.A.; Plummer, T.W.; Bose, R. A GIS-based approach to documenting large canid damage to bones. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2014, 409, 57–71. [Google Scholar] [CrossRef]
- Parkinson, J.A.; Plummer, T.W.; Harstone-Rose, A. Characterizing felid tooth marking and gross bone damage patterns using GIS image analysis: An experimental feeding study with large felids. J. Hum. Evol. 2015, 80, 114–134. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.A.; Plummer, T.W.; Oliver, J.S.; Bishop, L.C. Meat on the menu: GIS spatial distribution analysis of bone surface damage indicates that Oldowan hominins at Kanjera South, Kenya had early access to carcasses. Quat. Sci. Rev. 2022, 277, 107314. [Google Scholar] [CrossRef]
- Stavrova, T.; Borel, A.; Daujeard, C.; Vettese, D. A GIS based approach to long bone breakage patterns derived from marrow extraction. PLoS ONE 2019, 14, e0216733. [Google Scholar] [CrossRef] [PubMed]
- Marean, C.W.; Abe, Y.; Nilssen, P.J.; Stone, E.C. Estimating the minimum number of skeletal elements (MNE) in Zooarchaeology: A review and a new Image-analysis GIS approach. Am. Antiq. 2001, 66, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Rodrigo, M.; Gidna, A.; Baquedano, E.; Cobo-Sánchez, L.; Mora, R.; Courtenay, L.A.; González-Aguilera, D.; Maté-González, M.A.; Prieto-Herráez, D. A 3D taphonomic model of long bone modification by lions in medium-sized ungulate carcasses. Sci. Rep. 2021, 11, 4944. [Google Scholar] [CrossRef] [PubMed]
- Lyman, R.L. Quantitative Paleozoology, 1st ed.; Cambridge University Press: Cambridge, UK, 2008; p. 348. [Google Scholar]
- Discamps, E. TIPZOO: A Touchscreen Interface for Palaeolithic Zooarchaeology. Towards making data entry and analysis easier, faster, and more reliable. Peer Community J. 2021, 1, e67. [Google Scholar] [CrossRef]
- ESRI. ArcGIS Desktop, Version 10.4; Environmental Systems Research Institute: Redlands, CA, USA, 1999. Available online: https://desktop.arcgis.com/en/arcmap/10.4/get-started/setup/arcgis-desktop-quick-start-guide.htm(accessed on 15 May 2022).
- Domínguez-Rodrigo, M.; Baquedano, E.; Organista, E.; Cobo-Sánchez, L.; Mabulla, A.; Maskara, V.; Gidna, A.; Pizarro-Monzo, M.; Aramendi, J.; Galán, A.B.; et al. Early Pleistocene faunivorous hominins were not kleptoparasitic, and this impacted the evolution of human anatomy and socio-ecology. Sci. Rep. 2021, 11, 16135. [Google Scholar] [CrossRef]
- Pizarro-Monzo, M.; Prendergast, M.E.; Gidna, A.; Baquedano, E.; Mora, R.; González-Aguilera, D.; Maté-González, M.A.; Domínguez-Rodrigo, M. Do human butchery patterns exist? A study of the interaction of randomness and channelling in the distribution of cut marks on long bones. J. R. Soc. Interface 2021, 18, 20200958. [Google Scholar] [CrossRef]
- Brugal, J.; Boudadi-Maligne, M. Quaternary small to large canids in Europe: Taxonomic status and biochronological contribution. Quat. Int. 2011, 243, 171–182. [Google Scholar] [CrossRef]
- Campmas, É.; Beauval, C. Consommation osseuse des carnivores, résultats del’étude de l’exploitation de carcasses de boeufs (Bos taurus) par des loups captifs. Ann. Paléontologie 2008, 94, 167–186. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Maté-González, M.A.; Vázquez-Rodríguez, J.M.; Fernández-Fernández, M.; González-Aguilera, D. The effects of prey size on carnivore tooth mark morphologies on bone; the case study of Canis lupus signatus. Hist. Biol. 2020, 33, 2760–2772. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Herranz-Rodrigo, D.; Yravedra, J.; Vázquez-Rodríguez, J.M.; Huguet, R.; Barja, I.; Maté-González, M.A.; Fernández-Fernández, M.; Muñoz-Nieto, A.L.; González-Aguilera, D. 3D Insights into the Effects of Captivity on Wolf Mastication and Their Tooth Marks; Implications in Ecological Studies of Both the Past and Present. Animals 2021, 11, 2323. [Google Scholar] [CrossRef] [PubMed]
- Castel, J.-C. L’influence des canidés sur la formation des ensembles archéologiques: Caractérisation des destructions dues au loup. Rev. Paléobiologie 2004, 23, 675–693. [Google Scholar]
- Esteban-Nadal, M.; Cáceres, I.; Fosse, P. Characterization of a current coprogenic sample originated by Canis lupus as a tool for identifying a taphonomic agent. J. Archaeol. Sci. 2010, 37, 2959–2970. [Google Scholar] [CrossRef]
- Fosse, P.; Laudet, F.; Selva, N.; Wajrak, A. Premières observations néotaphonomiques sur des assemblages osseux de Bialowieza (N.E. Pologne): Intérêt pour les gisements pléistocènes d’Europe. Paléo 2004, 16, 91–116. [Google Scholar]
- Fosse, P.; Wajrak, A.; Fourvel, J.B.; Madelaine, S.; Esteban, M.; Cáceres, I.; Yravedra, J.; Brugal, J.P.; Prucca, A.; Haynes, G. Bone Modification by Modern Wolf (Canis lupus): A Taphonomic Study From their Natural Feeding Places. J. Taphon. 2012, 10, 197–217. [Google Scholar]
- Haynes, G. Utilization and skeletal disturbances of North American prey carcasses. Arctic 1982, 35, 266–281. [Google Scholar] [CrossRef]
- Sala, N.; Arsuaga, J.L.; Haynes, G. Taphonomic comparison of bone modifications caused by wild and captive wolves Canis lupus. Quat. Int. 2014, 330, 126–135. [Google Scholar] [CrossRef]
- Stiner, M.C. Comparative ecology and taphonomy of spotted hyenas, humans, and wolves in Pleistocene Italy. Rev. Paléobiologie 2004, 23, 771–785. [Google Scholar]
- Yravedra, J.; Lagos, L.; Bárcena, F. A taphonomic study of wild wolf (Canis lupus) modification of horse bones in northwestern Spain. J. Taphon. 2011, 9, 37–65. [Google Scholar]
- Yravedra, J.; Lagos, L.; Bárcena, F. The Wild Wolf (Canis lupus) as a Dispersal Agent of Animal Carcasses in Northwestern Spain. J. Taphon. 2012, 10, 227–248. [Google Scholar]
- Yravedra, J.; Maté-González, M.A.; Courtenay, L.A.; González-Aguilera, D.; Fernández-Fernández, M. The use of canid tooth marks on bone for the identification of livestock predation. Sci. Rep. 2019, 9, 16301. [Google Scholar] [CrossRef] [Green Version]
- Yravedra, J.; Herranz-Rodrigo, D.; Mendoza, C.; Aragón-Poza, P.; Courtenay, L.A. The use of tooth marks for new research into identifying and understanding the first domestic dogs in Palaeolithic populations. J. Archaeol. Sci. Rep. 2021, 40B, 103252. [Google Scholar] [CrossRef]
- Gidna, A.; Yravedra, J.; Domínguez-Rodrigo, M. A cautionary note on the use of captive carnivores to model wild predator behavior: A comparison of bone modification patterns on long bones by captive and wild lions. J. Archaeol. Sci. 2013, 40, 1903–1910. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.r-project.org/ (accessed on 15 May 2022).
- Courtenay, L.A.; Aramendi, J. IkhnosToolBox: R Tool Box for the Ikhnos Taphonomic Software, Version 1.0.0; GitHub, Inc.: San Francisco, CA, USA, 2022. Available online: https://github.com/TIDOP-USAL/IkhnosToolBox(accessed on 1 June 2022).
- Bunn, H.T. Meat-Eating and Human Evolution: Studies on the Diet and Subsistence Patterns of Plio-Pleistocene Hominids in East Africa. Ph.D. Dissertation, University of California, Berkeley, CA, USA, 1982. [Google Scholar]
- Ripley, B.D. The second-order analysis of stationary point processes. J. Appl. Probab. 1976, 13, 255–266. [Google Scholar] [CrossRef]
- Ripley, B.D. Modelling spatial patterns (with discussion). J. R. Stat. Soc. 1977, 39, 172–212. [Google Scholar]
- Ripley, B.D. Statistical Inference for Spatial Processes, Reprint ed.; Cambridge University Press: Cambridge, UK, 1991; p. 162. [Google Scholar]
- Grinsted, A.; Moore, J.C.; Jevrejeva, S. Application of the cross wavelet transform and wavelet coherence to geophysical time series. Nonlinear Process. Geophys. 2004, 11, 561–566. [Google Scholar] [CrossRef]
- Hinton, G.E.; Roweis, S.T. Stochastic Neighbor Embedding. Adv. Neural Inf. Process. Syst. 2002, 15, 857–864. [Google Scholar]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996. [Google Scholar]
- Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. Density-based clustering in spatial databases: The algorithm GDBSCAN and its applications. Data Min. Knowl. Discov. 1998, 2, 169–194. [Google Scholar] [CrossRef]
- Baddeley, A.J.; Moyeed, R.A.; Howard, C.V.; Boyde, A. Analysis of a three-dimensional point pattern with replication. Appl. Stat. 1993, 42, 641–668. [Google Scholar] [CrossRef]
- Baddeley, A.J.; Gill, R.D. Kaplan-Meier estimators of interpoint distance distributions for spatial point processes. Ann. Stat. 1997, 25, 263–292. [Google Scholar] [CrossRef]
- Chiu, S.N.; Stoyan, D. Estimators of distance distributions for spatial patterns. Stat. Neerl. 1998, 52, 239–246. [Google Scholar] [CrossRef]
- Cressie, N.A.C. Statistics for Spatial Data. Revised Edition, 2nd ed.; Wiley-Blackwell: New York, NY, USA, 1993; p. 928. [Google Scholar]
- Hanisch, K.H. Some remarks on estimators of the distribution function of nearest neighbour distance in stationary spatial point patterns. Math. Oper. Stat. 1984, 15, 409–412. [Google Scholar]
- Ohser, J. On estimators for the reduced second moment measure of point processes. Math. Oper. Stat. 1983, 14, 63–71. [Google Scholar] [CrossRef]
- Baddeley, A. , Rubak, E., Turner, R. Spatial Point Patterns: Methodology and Applications with R, 1st ed.; Chapman and Hall: Boca Raton, FL, USA, 2015; p. 828. [Google Scholar]
- Cazelles, B.; Chavez, M.; Berteaux, D.; Menard, F.; Vik, J.O.; Jenouvrier, S.; Stenseth, N.C. Wavelet analysis of ecological time series. Oecologia 2008, 156, 287–304. [Google Scholar] [CrossRef]
- Chavez, M.; Cazelles, B. Detecting dynamic spatial correlation patterns with generalized wavelet coherence and non-stationary surrogate data. Sci. Rep. 2019, 9, 7389. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; San Liang, X.; Weisberg, R.H. Rectification of the Bias in the Wavelet Power Spectrum. J. Atmos. Ocean. Technol. 2007, 24, 2093–2102. [Google Scholar] [CrossRef]
- Nason, G. Wavelet Methods in Statistics with R, 1st ed.; Springer: New York, NY, USA, 2008; p. 272. [Google Scholar]
- Torrence, C.; Compo, G.P. A Practical Guide to Wavelet Analysis. Bull. Am. Meteorol. Soc. 1998, 79, 61–78. [Google Scholar] [CrossRef]
- Veleda, D.; Montagne, R.; Araujo, M. Cross-Wavelet Bias Corrected by Normalizing Scales. J. Atmos. Ocean. Technol. 2012, 29, 1401–1408. [Google Scholar] [CrossRef]
- Mardia, K.V. Statistics of Directional Data. J. R. Stat. Soc. 1975, 37, 349–393. [Google Scholar] [CrossRef]
- Watson, G.S. Analysis of dispersion on a sphere. Geophys. J. Int. 1956, 7, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Edelsbrunner, H.; Kirkpatrick, D.G.; Seidel, R. On the shape of a set of points in the plane. IEEE Trans. Inf. Theory 1983, 29, 551–559. [Google Scholar] [CrossRef] [Green Version]
- Jammalamadaka, S.R.; SenGupta, A. Topics in Circular Statistics, 1st ed.; World Scientific Press: Singapore, 2001; p. 336. [Google Scholar]
- Wheeler, S.; Watson, G.S. A distribution-free two-sample test on the circle. Biometrika 1964, 51, 256–257. [Google Scholar] [CrossRef]
- pValueRobust: A Library for Robust Statistical Measures and the Evaluation of p-Values. Version 0.1.0. Available online: https://github.com/LACourtenay/pValueRobust (accessed on 15 June 2022).
- Colquhoun, D. The False Positive Risk: A Proposal Concerning What to do about p-Values. Am. Stat. 2019, 73 (Suppl. S1), 192–201. [Google Scholar] [CrossRef] [Green Version]
- Domínguez-Rodrigo, M. Hunting and scavenging by early humans, the state of the debate. J. World Prehistory 2002, 16, 1–54. [Google Scholar] [CrossRef]
- R Package Biwavelet: Conduct Univariate and Bivariate Wavelet Analyses. Available online: https://github.com/tgouhier/biwavelet (accessed on 1 June 2022).
- R Package ‘Circular’: Circular Statistics (Version 0.4-94). Available online: https://r-forge.r-project.org/projects/circular/ (accessed on 1 July 2022).
- Rtsne: T-Distributed Stochastic Neighbor Embedding Using a Barnes-Hut Implementation. Available online: https://github.com/jkrijthe/Rtsne (accessed on 1 June 2022).
- Domínguez-Rodrigo, M.; Yravedra, J.; Organista, E.; Gidna, A.; Fourvel, J.B.; Baquedano, E. A new methodological approach to the taphonomic study of paleontological and archaeological faunal assemblages: A preliminary case study from Olduvai Gorge (Tanzania). J. Archaeol. Sci. 2015, 59, 35–53. [Google Scholar] [CrossRef]
- Haynes, G. A Guide for Differentiating Mammalian Carnivore Taxa Responsible for Gnaw Damage to herbivore Limb Bones. Paleobiology 1983, 9, 164–172. [Google Scholar] [CrossRef]
- Marean, C.W.; Frey, C.J. Animal bones from caves to cities: Reverse utility curves as methodological artifacts. Am. Antiq. 1997, 62, 698–711. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Gidna, A.O.; Yravedra, J.; Musiba, C. A comparative neo-taphonomic study of felids, hyaenids and canids: An analogical framework based on long bone modification patterns. J. Taphon. 2012, 10, 121–169. [Google Scholar]
- Organista, E.; Pernas-Hernández, M.; Gidna, A.; Yravedra, J.; Domínguez-Rodrigo, M. An experimental lion-to-hammerstone model and its relevance to understand hominin-carnivore interactions in the archaeological record. J. Archaeol. Sci. 2016, 66, 69–77. [Google Scholar] [CrossRef]
- Kuhn, B.F.; Berger, L.R.; Skinner, J.D. Variation in tooth mark frequencies on long bones from the assemblages of all three extant bone-collecting hyaenids. J. Archaeol. Sci. 2009, 36, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Faith, J.T. Sources of variation in carnivore tooth-mark frequencies in a modern spotted hyena (Crocuta crocuta) den assemblage, Amboseli Park, Kenya. J. Archaeol. Sci. 2007, 34, 1601–1609. [Google Scholar] [CrossRef]
- Ruckthongsook, W.; Tiowari, C.; Oppong, J.; Natesan, P. Evaluation of threshold selection methods for adaptive kernel density estimation in disease mapping. Int. J. Health Geogr. 2018, 8, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Bunn, H.T.; Bartram, L.E.; Kroll, L.M. Variability in bone assemblage formation from Hadza hunting, scavenging, and carcass processing. J. Anthropol. Archaeol. 1988, 7, 412–457. [Google Scholar]



| Software | Bone Elements | Data Collection | Statistics | Access |
|---|---|---|---|---|
| ArcGis 1 (ArcMap) 1 | All types | •Bone survivorship is registered via digital drawing over photographic 2D templates of complete bones •BSMs are plotted on separated rasterized 2D bone templates for anterior, posterior, mesial, and lateral views of each element | Spatial analyses including nearest neighbor distance, kernel density estimation | License fee |
| TIPZOO 2 | All types | •Bone survivorship and BSM documented through codes according to skeletal element, segment and orientation | Data export for statistical analysis in separate software tools, not suited for point-based spatial statistics | Open Access |
| Ikhnos | Long bones | •Bone survivorship is registered via digital polygonal cropping on the 3D point mesh and the calculation of heatmaps •BSMs are documented through the selection of landmarks (x,y,z) on the 3D point mesh of each element | Integrated R library IkhnosToolBox that includes 3D spatial and multivariate statistical analyses, tests for point patterning identification and visualization, and circular analyses for linear BSMs | Open Access |
| Sample | Femur | Tibia | ||||||
|---|---|---|---|---|---|---|---|---|
| Right (C = 4, W = 6) | Left (C = 8, W = 3) | Right (C = 14, W = 1) | Left (C = 15, W = 1) | |||||
| pits | scores | pits | scores | pits | scores | pits | scores | |
| Wild wolves (W) | 22 | 29 | 26 | 32 | 15 | 11 | 10 | 16 |
| Captive wolves (C) | 130 | 39 | 240 | 45 | 769 | 363 | 431 | 188 |
| Statistics | Right Femur | Left Femur | Right Tibia | Left Tibia | ||||
|---|---|---|---|---|---|---|---|---|
| Test | Captive | Wild | Captive | Wild | Captive | Wild | Captive | Wild |
| Skewness | −0.8651 | 0.3926 | 0.1974 | 0.9063 | 0.1824 | −1.0769 | −0.1009 | −1.7587 |
| Kurtosis | −0.1101 | −0.6033 | 1.1583 | 0.9208 | 1.816 | 3.0222 | 3.6393 | 0.7149 |
| Circular variance | 0.2741 | 0.2523 | 0.1884 | 0.2535 | 0.1671 | 0.0827 | 0.1266 | 0.0681 |
| Circular dispersion | 0.6676 | 0.6451 | 0.3982 | 0.5497 | 0.3373 | 0.1609 | 0.2357 | 0.1392 |
| Central orientation radians | −3.13/0.01 | 3.06/−0.08 | −0.05/3.09 | 0.36/−2.78 | 0.06/−3.08 | −0.04/3.1 | −0.02/3.12 | 0.7/−2.44 |
| Central orientation degrees | −179.3/0.7 | 175.2/−4.8 | −3.1/176.9 | 20.9/−159.1 | 3.6/−176.4 | −2.1/177.9 | −1.2/178.8 | 39.9/−140.1 |
| Preferential orientation | 0.7258 (1.0 × 10−9) | 0.7477 (3.5 × 10−8) | 0.8116 (6.9 × 10−13) | 0.7465 (1.0 × 10−8) | 0.8329 (4.4 × 10−110) | 0.9173 (0) | 0.8734 (5.1 × 10−63) | 0.9319 (5.0 × 10−7) |
| Orientation comparison | 6.7226 × 10−31 (p = 1) | 4.8088 × 10−31 (p = 1) | 2.3845 × 10−30 (p = 1) | 1.974 × 10−31 (p = 1) | ||||
| Captive Femora: 6.1859 × 10−31 (p = 1) Wild Femora: 4.8418 × 10−31 (p = 1) | Captive Tibiae: 3.3887 × 10−30 (p = 1) Wild Tibiae: 4.8725 × 10−31 (p = 1) | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora, R.; Aramendi, J.; Courtenay, L.A.; González-Aguilera, D.; Yravedra, J.; Maté-González, M.Á.; Prieto-Herráez, D.; Vázquez-Rodríguez, J.M.; Barja, I. Ikhnos: A Novel Software to Register and Analyze Bone Surface Modifications Based on Three-Dimensional Documentation. Animals 2022, 12, 2861. https://doi.org/10.3390/ani12202861
Mora R, Aramendi J, Courtenay LA, González-Aguilera D, Yravedra J, Maté-González MÁ, Prieto-Herráez D, Vázquez-Rodríguez JM, Barja I. Ikhnos: A Novel Software to Register and Analyze Bone Surface Modifications Based on Three-Dimensional Documentation. Animals. 2022; 12(20):2861. https://doi.org/10.3390/ani12202861
Chicago/Turabian StyleMora, Rocío, Julia Aramendi, Lloyd A. Courtenay, Diego González-Aguilera, José Yravedra, Miguel Ángel Maté-González, Diego Prieto-Herráez, José Mª Vázquez-Rodríguez, and Isabel Barja. 2022. "Ikhnos: A Novel Software to Register and Analyze Bone Surface Modifications Based on Three-Dimensional Documentation" Animals 12, no. 20: 2861. https://doi.org/10.3390/ani12202861







