Phylogenetic Relationships of the Strongyloid Nematodes of Australasian Marsupials Based on Mitochondrial Protein Sequences
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sequencing and Gene Annotation
2.3. Sequence Comparison and Phylogenetic Analyses
3. Results
3.1. Nucleotide and Amino Acid Sequence Comparisons
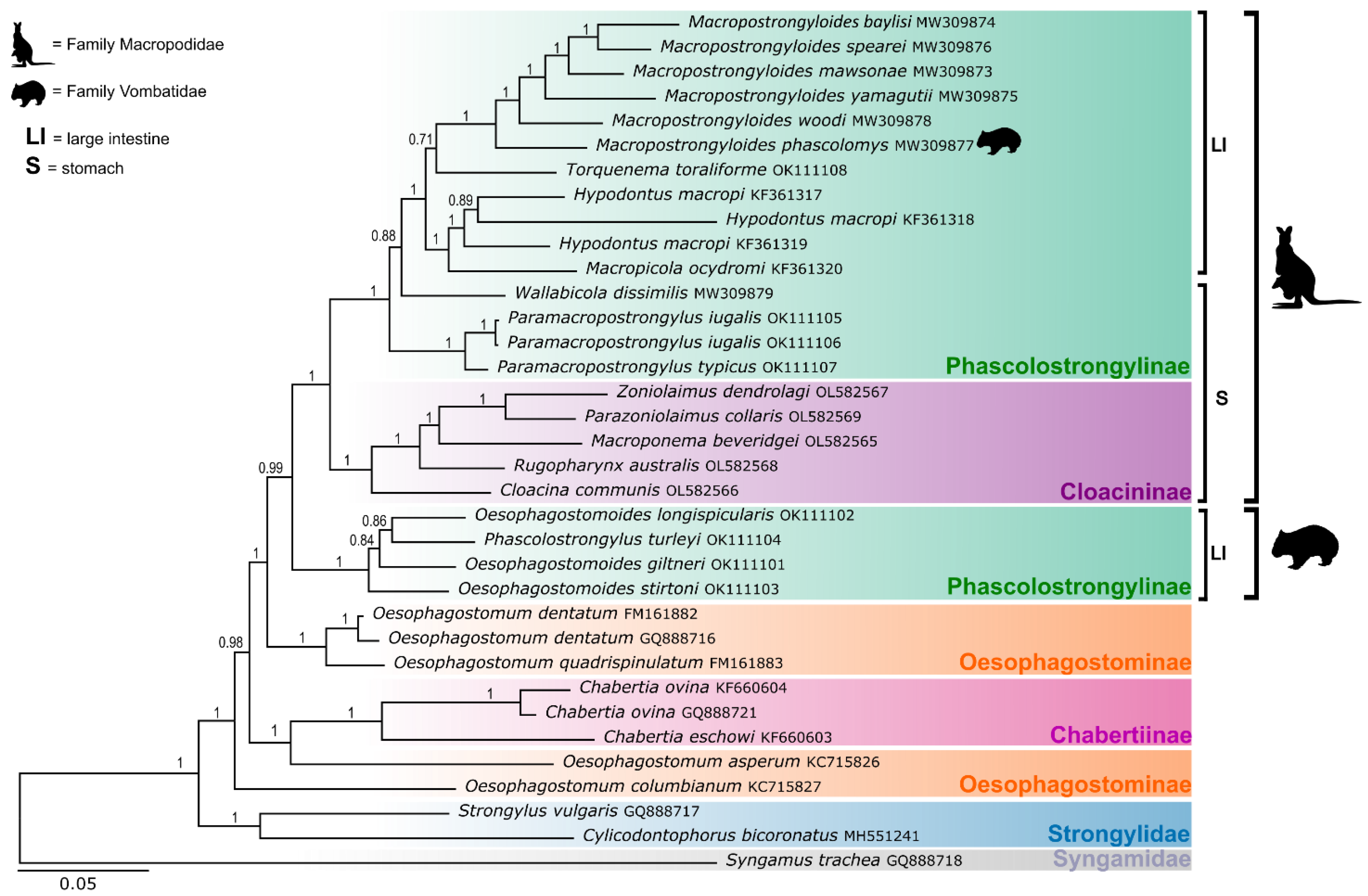
3.2. Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spratt, D.M.; Beveridge, I. Helminth parasites of Australasian monotremes and marsupials. Zootaxa 2016, 4123, 1–198. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, I. The systematic status of Australian Strongyloidea (Nematoda). Bull. Mus. Natl. Hist. Nat. 1987, 9, 107–126. [Google Scholar]
- Beveridge, I.; Smales, L. Review of the Cloacininae Stossich (Nemata: Strongyloidea) from Australasian marsupials (Marsupialia: Macropodoidea). MANTER J. Parasit. Biodivers. 2022, 21, 1–197. [Google Scholar] [CrossRef]
- Beveridge, I.; Jabbar, A.; Koehler, A.V.; Sukee, T. A morphological and molecular phylogenetic analysis of relationships between genera of the nematode sub-family Cloacininae (Stossich) (Strongyloidea: Chabertiidae) parasitic in kangaroos, wallabies and rat-kangaroos (Marsupialia: Macropodoidea). Zootaxa 2020, 4851, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Sukee, T.; Beveridge, I.; Koehler, A.V.; Hall, R.; Gasser, R.B.; Jabbar, A. Phylogenetic relationships of the nematode subfamily Phascolostrongylinae from macropodid and vombatid marsupials inferred using mitochondrial protein sequence data. Parasit. Vectors 2021, 14, 523. [Google Scholar] [CrossRef]
- Sukee, T.; Beveridge, I.; Sabir, A.J.; Jabbar, A. Phylogenetic relationships within the nematode subfamily Phascolostrongylinae (Nematoda: Strongyloidea) from Australian macropodid and vombatid marsupials. Microorganisms 2021, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Chilton, N.B.; Gasser, R.B.; Beveridge, I. Phylogenetic relationships of Australian strongyloid nematodes inferred from ribosomal DNA sequence data. Int. J. Parasitol. 1997, 27, 1481–1494. [Google Scholar] [CrossRef]
- Jabbar, A.; Beveridge, I.; Namitha, M.; Chilton, N.B.; Littlewood, T.J.; Jex, A.R.; Gasser, R.B. Analyses of mitochondrial amino acid sequence datasets support the proposal that specimens of Hypodontus macropi from three species of macropodid hosts represent distinct species. BMC Evol. Biol. 2013, 13, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukee, T.; Koehler, A.V.; Hall, R.; Beveridge, I.; Gasser, R.B.; Jabbar, A. Phylogenetic analysis of mitogenomic data sets resolves the relationship of seven Macropostrongyloides species from Australian macropodid and vombatid marsupials. Pathogens 2020, 9, 1042. [Google Scholar] [CrossRef]
- Lin, R.Q.; Liu, G.H.; Hu, M.; Song, H.Q.; Wu, X.Y.; Li, M.W.; Zhang, Y.; Zou, F.C.; Zhu, X.Q. Oesophagostomum dentatum and Oesophagostomum quadrispinulatum: Characterization of the complete mitochondrial genome sequences of the two pig nodule worms. Exp. Parasitol. 2012, 131, 1–7. [Google Scholar] [CrossRef]
- Jex, A.R.; Hall, R.S.; Littlewood, T.J.; Gasser, R.B. An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res. 2010, 38, 522–533. [Google Scholar] [CrossRef]
- Zhao, G.H.; Zhao, L.; Song, H.-Q.; Zhao, G.-H.; Cai, J.-Z.; Zhao, Q.; Zhu, X.-Q. The complete mitochondrial genomes of Oesophagostomum asperum and Oesophagostomum columbianum in small ruminants. Infect. Genet. Evol. 2013, 19, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-H.; Zhao, L.; Song, H.-Q.; Zhao, G.-H.; Cai, J.-Z.; Zhao, Q.; Zhu, X.-Q. Chabertia erschowi (Nematoda) is a distinct species based on nuclear ribosomal DNA sequences and mitochondrial DNA sequences. Parasit. Vectors 2014, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wang, X.X.; Ma, X.X.; Zhang, Z.H.; Lan, Z.; Qiu, Y.Y.; Wang, S.; Song, M.X.; Wang, C.R. Characterization of the complete mitochondrial genomes of Coronocyclus labiatus and Cylicodontophorus bicoronatus: Comparison with Strongylidae species and phylogenetic implication. Vet. Parasitol. 2021, 290, 109359. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1. 3.1. 2009. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 18 October 2021).
- Lichtenfels, J.R. A conventional approach to a new classification of the Strongyloidea, nematode parasites of mammals. Am. Zool. 1979, 19, 1185–1194. [Google Scholar] [CrossRef]
- Lichtenfels, J.R. CIH Keys to the Nematode Parasites of Vertebrates—No. 7. Keys to the Genera of the Superfamily Strongyloidea; Commonwealth Agriculture Bureaux: Farnham Royal, Bucks, UK, 1980; pp. 1–41. [Google Scholar]
- Sukee, T.; Beveridge, I.; Jabbar, A. Torquenema n. g., Wallabicola n. g., and Macropostrongyloides phascolomys n. sp.: New genera and a new species of nematode (Strongylida: Phascolostrongylinae) parasitic in Australian macropodid and vombatid marsupials. Animals 2021, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, I. New species of parasitic nematodes of the genus Cloacina (Nematoda: Strongyloidea) from the banded hare wallaby, Lagostrophus fasciatus (Marsupialia: Macropodidae). Trans. R. Soc. S. Aust. 2014, 138, 249–256. [Google Scholar] [CrossRef]
- Beveridge, I.; Spratt, D.M. The helminth fauna of Australasian marsupials: Origins and evolutionary biology. Adv. Parasitol. 1996, 37, 135–254. [Google Scholar]
- Beveridge, I. Evolution of the strongyloid nematodes of Australian marsupials. Bull. Mus. Nat. Hist. Nat. Paris Sér. A 1982, 9, 107–126. [Google Scholar]
- Beck, R.M.; Louys, J.; Brewer, P.; Archer, M.; Black, K.H.; Tedford, R.H. A new family of diprotodontian marsupials from the latest Oligocene of Australia and the evolution of wombats, koalas, and their relatives (Vombatiformes). Sci. Rep. 2020, 10, 9741. [Google Scholar] [CrossRef]
- Meredith, R.W.; Westerman, M.; Case, J.A.; Springer, M.S. A phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J. Mamm. Evol. 2008, 15, 1–36. [Google Scholar] [CrossRef]
- Hardman, L.M.; Haukisalmi, V.; Beveridge, I. Phylogenetic relationships of the anoplocephaline cestodes of Australasian marsupials and resurrection of the genus Wallabicestus Schmidt, 1975. Syst. Parasitol. 2012, 82, 49–63. [Google Scholar] [CrossRef]
- Van Dyck, S.; Strahan, R. Mammals of Australia, 3rd ed.; Reed New Holland: Sydney, Australia, 2008; pp. 1–888. [Google Scholar]
- Wood, J.R.; Wilmshurst, J.M.; Rawlence, N.J.; Bonner, K.I.; Worthy, T.H.; Kinsella, J.M.; Cooper, A.A. Megafauna’s microfauna: Gastrointestinal parasites of New Zealand’s extinct moa (Aves: Dinornithiformes). PLoS ONE 2013, 8, e57315. [Google Scholar]
- Hume, I.D. Digestive Physiology and Nutrition of Marsupials, in Monographs on Marsupial Biology; Cambridge University Press: Cambridge, UK, 1982; p. 265. [Google Scholar]
- Kapli, P.; Yang, Z.; Telford, Z. Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 2020, 21, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Mar, J.C.; Harlow, T.J.; Ragan, M.A. Bayesian and maximum likelihood phylogenetic analyses of protein sequence data under relative branch-length differences and model violation. BMC Evol. Biol. 2005, 5, 8. [Google Scholar] [CrossRef] [PubMed]

| Family or Subfamily | Species | Host Species | Collection Locality | GenBank Accession Mo. | Reference |
|---|---|---|---|---|---|
| Subfamily Cloacininae | Cloacina communis | Osphranter robustus | Menzies, WA, Australia | OL582566 | this study |
| Macroponema beveridgei | Osphranter robustus | Hillgrove Stn, Charters Towers, Qld, Australia | OL582565 | this study | |
| Parazoniolaimus collaris | Wallabia bicolor | The Gurdies, Vic, Australia | OL582569 | this study | |
| Rugopharynx australis | Osphranter rufus | Wallerberdina Stn, Port Augusta, SA, Australia | OL582568 | this study | |
| Zoniolaimus dendrolagi | Dendrolagus lumholtzi | East Beatrice, Qld, Australia | OL582567 | this study | |
| Subfamily Phascolostrongylinae | Oesophagostomoides giltneri | Vombatus ursinus | Flowerdale, Vic, Australia | OK111101 | [5] |
| Oesophagostomoides longispicularis | Vombatus ursinus | Gippsland, Vic, Australia | OK111102 | [5] | |
| Oesophagostomoides stirtoni | Lasiorhinus latifrons | Swan Reach, SA, Australia | OK111103 | [5] | |
| Paramacropostrongylus iugalis | Macropus giganteus | Miles, Qld, Australia | OK111105 | [5] | |
| Paramacropostrongylus iugalis | Macropus giganteus | Charters Towers, Qld, Australia | OK111106 | [5] | |
| Paramacropostrongylus typicus | Macropus fuliginosus | Nyngan, NSW, Australia | OK111107 | [5] | |
| Phascolostrongylus turleyi | Vombatus ursinus | Flowerdale, Vic, Australia | OK111104 | [5] | |
| Torquenema toraliforme | Macropus giganteus | Research, Vic, Australia | OK111108 | [5] | |
| Macropostrongyloides mawsonae | Macropus giganteus | Heathcote, Vic, Australia | MW309873 | [9] | |
| Macropostrongyloides baylisi | Osphranter robustus | Cloncurry, Qld, Australia | MW309874 | [9] | |
| Macropostrongyloides yamagutii | Macropus fuliginosus | Hattah Lakes, Vic, Australia | MW309875 | [9] | |
| Macropostrongyloides spearei | Osphranter robustus | Kalgoorlie, WA, Australia | MW309876 | [9] | |
| Macropostrongyloides phascolomys | Vombatus ursinus | Flowerdale, Vic, Australia | MW309877 | [9] | |
| Macropostrongyloides woodi | Osphranter rufus | Kalgoorlie, WA, Australia | MW309878 | [9] | |
| Wallabicola dissimilis | Wallabia bicolor | Kamarooka, Vic, Australia | MW309879 | [9] | |
| Hypodontus macropi | Wallabia bicolor | Hall’s Gap, Vic, Australia | KF361317 | [8] | |
| Hypodontus macropi | Thylogale billardierii | Launceston, Tas, Australia | KF361318 | [8] | |
| Hypodontus macropi | Macropus robustus | Barcaldine, Qld, Australia | KF361319 | [8] | |
| Macropicola ocydromi | Macropus fuliginosus | Waroona, WA, Australia | KF361320 | [8] | |
| Subfamily Oesophagostominae | Oesophagostomum dentatum | Sus scrofa domestica | Chongqing, China | FM161882 | [10] |
| Oesophagostomum quadrispinulatum | Sus scrofa domestica | Chongqing, China | FM161883 | [10] | |
| Oesophagostomum dentatum | Sus scrofa domestica | Werribee, Vic, Australia | GQ888716 | [11] | |
| Oesophagostomum asperum | Capra hircus | Shaanxi Province, China | KC715826 | [12] | |
| Oesophagostomum columbianum | Ovis aries | Heilongjiang Province, China | KC715827 | [12] | |
| Subfamily Chabertiinae | Chabertia ovina | Ovis aries | Werribee, Vic, Australia | GQ888721 | [11] |
| Chabertia ovina | Capra hircus | Shaanxi Province, China | KF660604 | [13] | |
| Chabertia ershowi | Bos grunniens | Qinghai Province, China | KF660603 | [13] | |
| Family Strongylidae | Cylicodontophorus bicoronatus | Equus caballus | Heilongjiang Province, China | MH551241 | [14] |
| Strongylus vulgaris | Equus caballus | Werribee, Vic, Australia | GQ888717 | [11] | |
| Family Syngamidae | Syngamus trachea | Gymnorhina tibicen | Werribee, Vic, Australia | GQ888718 | [11] |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. C. communis 35T2 | 14.3 | 14.2 | 13.9 | 15.4 | 15.8 | 15.2 | 15.1 | 15.0 | 14.9 | 14.3 | 15.3 | 14.5 | 14.7 | 14.8 | 15.0 | 14.9 | 15.9 | |
| 2. Mac. beveridgei 21H1 | 7.9 | 13.6 | 12.8 | 14.4 | 15.8 | 15.5 | 15.7 | 15.3 | 14.9 | 14.1 | 15.6 | 14.8 | 15.5 | 15.4 | 15.3 | 15.0 | 16.0 | |
| 3. Pa. collaris YE8 | 8.3 | 7.7 | 12.9 | 13.1 | 15.7 | 15.4 | 15.4 | 15.4 | 14.6 | 14.4 | 15.6 | 14.9 | 15.2 | 15.4 | 15.3 | 14.9 | 15.7 | |
| 4. R. australis AC5 | 6.5 | 6.7 | 6.2 | 14.2 | 15.4 | 14.7 | 14.9 | 14.8 | 14.0 | 14.3 | 15.1 | 14.3 | 15.2 | 15.0 | 15.0 | 14.4 | 15.7 | |
| 5. Z. dendrolagi 48Z2 | 8.4 | 7.8 | 5.4 | 7.2 | 16.6 | 16.2 | 15.8 | 15.9 | 15.3 | 15.3 | 16.5 | 15.8 | 15.9 | 16.4 | 15.9 | 16.1 | 16.9 | |
| 6. Ma. baylisi 21v1 | 9.7 | 10.6 | 10.7 | 9.5 | 11.1 | 15.1 | 15.0 | 15.1 | 14.2 | 14.5 | 13.7 | 16.2 | 16.1 | 16.0 | 16.3 | 15.8 | 16.7 | |
| 7. H. macropi KF361317 | 8.4 | 8.9 | 9.6 | 8.2 | 9.8 | 8.4 | 13.6 | 14.8 | 13.8 | 14.0 | 14.2 | 15.3 | 15.6 | 15.8 | 15.9 | 15.3 | 16.0 | |
| 8. M. ocydromi KF361320 | 8.8 | 9.9 | 10.0 | 8.9 | 10.3 | 8.3 | 6.5 | 14.4 | 13.6 | 14.2 | 14.2 | 15.4 | 15.3 | 16.1 | 15.7 | 15.4 | 16.2 | |
| 9. W. dissimilis 10W9 | 7.9 | 8.8 | 9.5 | 8.2 | 9.5 | 8.5 | 6.5 | 7.2 | 13.6 | 13.3 | 14.3 | 15.2 | 15.3 | 15.8 | 15.5 | 14.9 | 16.0 | |
| 10. T. toraliforme YD5 | 8.4 | 9.5 | 9.5 | 8.3 | 10.1 | 7.9 | 6.9 | 7.1 | 6.6 | 12.9 | 13.5 | 14.7 | 15.4 | 15.1 | 15.2 | 14.7 | 15.8 | |
| 11. P. typicus 14B28 | 7.8 | 8.9 | 8.8 | 7.6 | 9.2 | 8.2 | 6.7 | 7.1 | 5.8 | 6.4 | 13.9 | 14.6 | 14.7 | 14.8 | 14.4 | 14.4 | 15.7 | |
| 12. Ma. phascolomys 41R1 | 9.1 | 10.5 | 10.4 | 9.3 | 10.8 | 7.0 | 7.5 | 7.5 | 7.4 | 7.2 | 7.4 | 15.7 | 15.6 | 15.8 | 15.8 | 15.6 | 16.7 | |
| 13. O. stirtoni 41W1 | 7.7 | 9.2 | 9.1 | 8.1 | 9.4 | 10.1 | 8.2 | 9.2 | 8.3 | 8.2 | 7.8 | 9.2 | 12.6 | 12.5 | 12.4 | 14.3 | 15.5 | |
| 14. O. giltneri 41Z1 | 8.3 | 9.5 | 9.4 | 8.6 | 9.7 | 10.1 | 8.5 | 9.4 | 8.4 | 8.8 | 8.0 | 9.7 | 4.9 | 12.4 | 12.4 | 14.5 | 16.2 | |
| 15. Ph. turleyi 42L2 | 8.2 | 9.9 | 9.9 | 8.6 | 9.9 | 10.4 | 9.3 | 9.8 | 8.8 | 9.0 | 8.4 | 9.8 | 4.7 | 4.9 | 12.4 | 14.5 | 16.1 | |
| 16. O. longispicularis 47F | 7.85 | 9.36 | 9.39 | 8.17 | 9.42 | 10.1 | 8.3 | 9.5 | 8.3 | 8.3 | 7.9 | 9.4 | 4.9 | 4.8 | 4.9 | 14.5 | 16.2 | |
| 17. Oe. dentatum FM161882 | 7.59 | 8.6 | 8.08 | 7.88 | 8.95 | 9.91 | 8.1 | 8.9 | 7.9 | 8.3 | 7.7 | 9.2 | 6.4 | 6.7 | 7.0 | 6.6 | 15.6 | |
| 18. Ch. ovina GQ888721 | 11.2 | 12 | 11.9 | 11.4 | 12.2 | 13 | 11.3 | 11.8 | 11.2 | 11.5 | 11.2 | 12.1 | 10.4 | 10.6 | 11.3 | 10.9 | 9.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sukee, T.; Beveridge, I.; Koehler, A.V.; Hall, R.S.; Gasser, R.B.; Jabbar, A. Phylogenetic Relationships of the Strongyloid Nematodes of Australasian Marsupials Based on Mitochondrial Protein Sequences. Animals 2022, 12, 2900. https://doi.org/10.3390/ani12212900
Sukee T, Beveridge I, Koehler AV, Hall RS, Gasser RB, Jabbar A. Phylogenetic Relationships of the Strongyloid Nematodes of Australasian Marsupials Based on Mitochondrial Protein Sequences. Animals. 2022; 12(21):2900. https://doi.org/10.3390/ani12212900
Chicago/Turabian StyleSukee, Tanapan, Ian Beveridge, Anson V. Koehler, Ross S. Hall, Robin B. Gasser, and Abdul Jabbar. 2022. "Phylogenetic Relationships of the Strongyloid Nematodes of Australasian Marsupials Based on Mitochondrial Protein Sequences" Animals 12, no. 21: 2900. https://doi.org/10.3390/ani12212900







