The Analysis of Transcriptomes and Microorganisms Reveals Differences between the Intestinal Segments of Guinea Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals Materials
2.3. Total RNA Extraction, Library Preparation and Sequencing
2.4. Quantification of PCGs and lncRNAs
2.5. Tissue Specific and Alternative Splicing Analysis
2.6. Differentially Expressed Genes Analysis and Functional Enrichments
2.7. Gene Set and Cell Composition Analysis
2.8. DNA Isolation, Library Preparation and Sequencing
2.9. Weighted Gene Correlation Network Analysis
3. Results
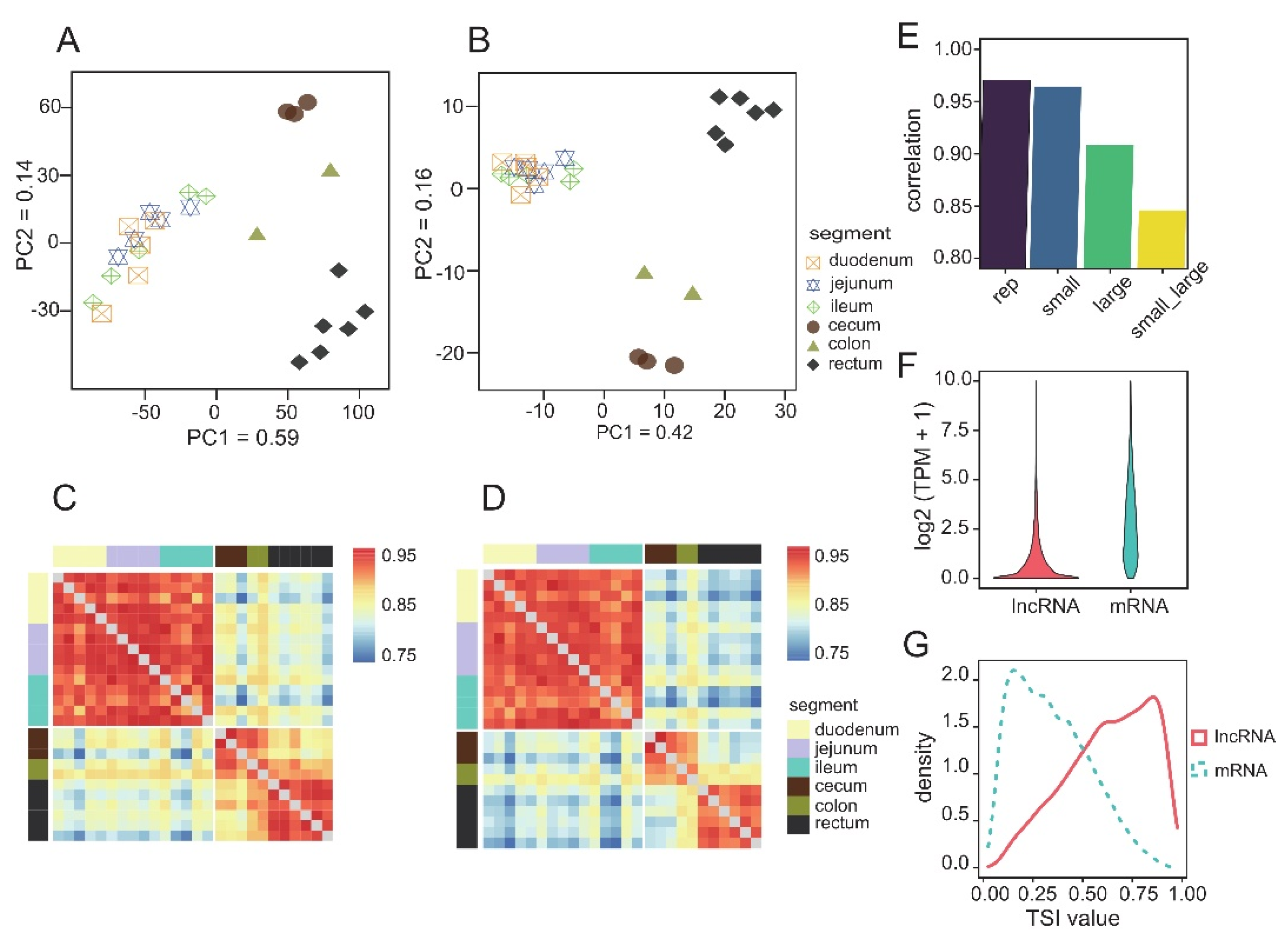
3.1. Comparison of the Transcription Profiles of the Different Intestinal Segments of Guinea Pigs
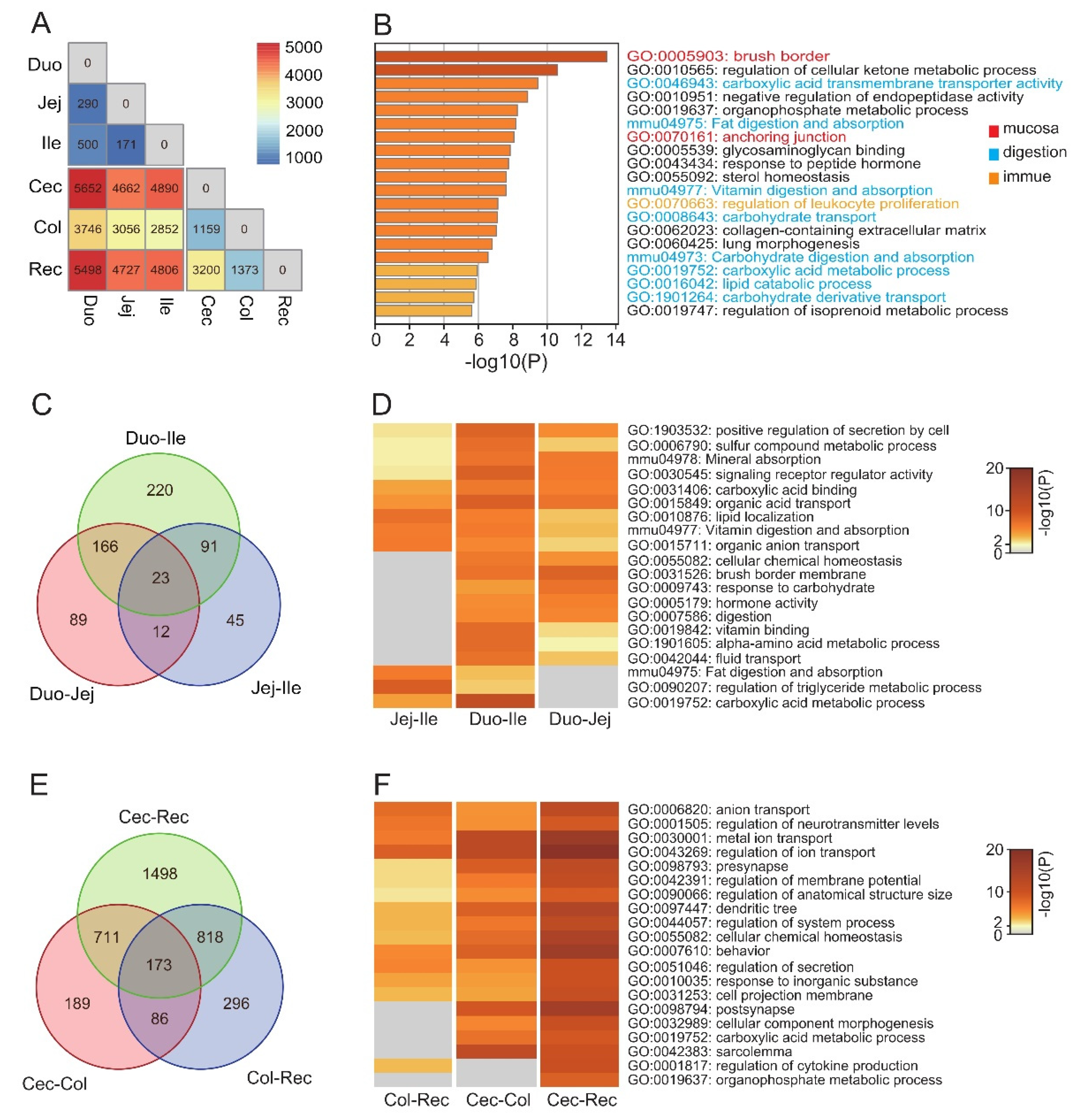
3.2. Functional Differences between Intestinal Segments
3.3. Distinct Expression Patterns of the Functional Gene set in the Intestines
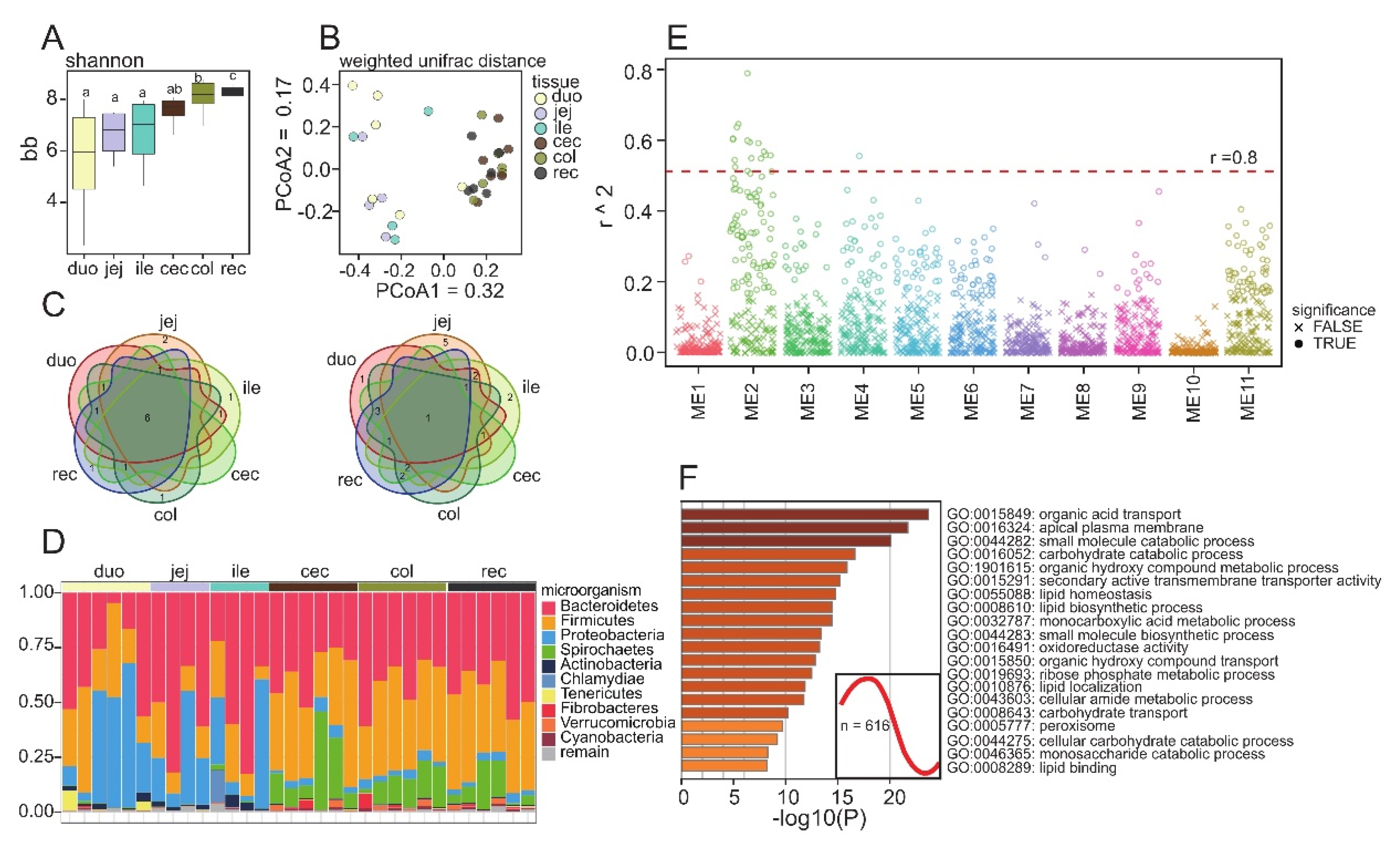
3.4. Microbial Composition of the Different Intestinal Segments of Guinea Pigs
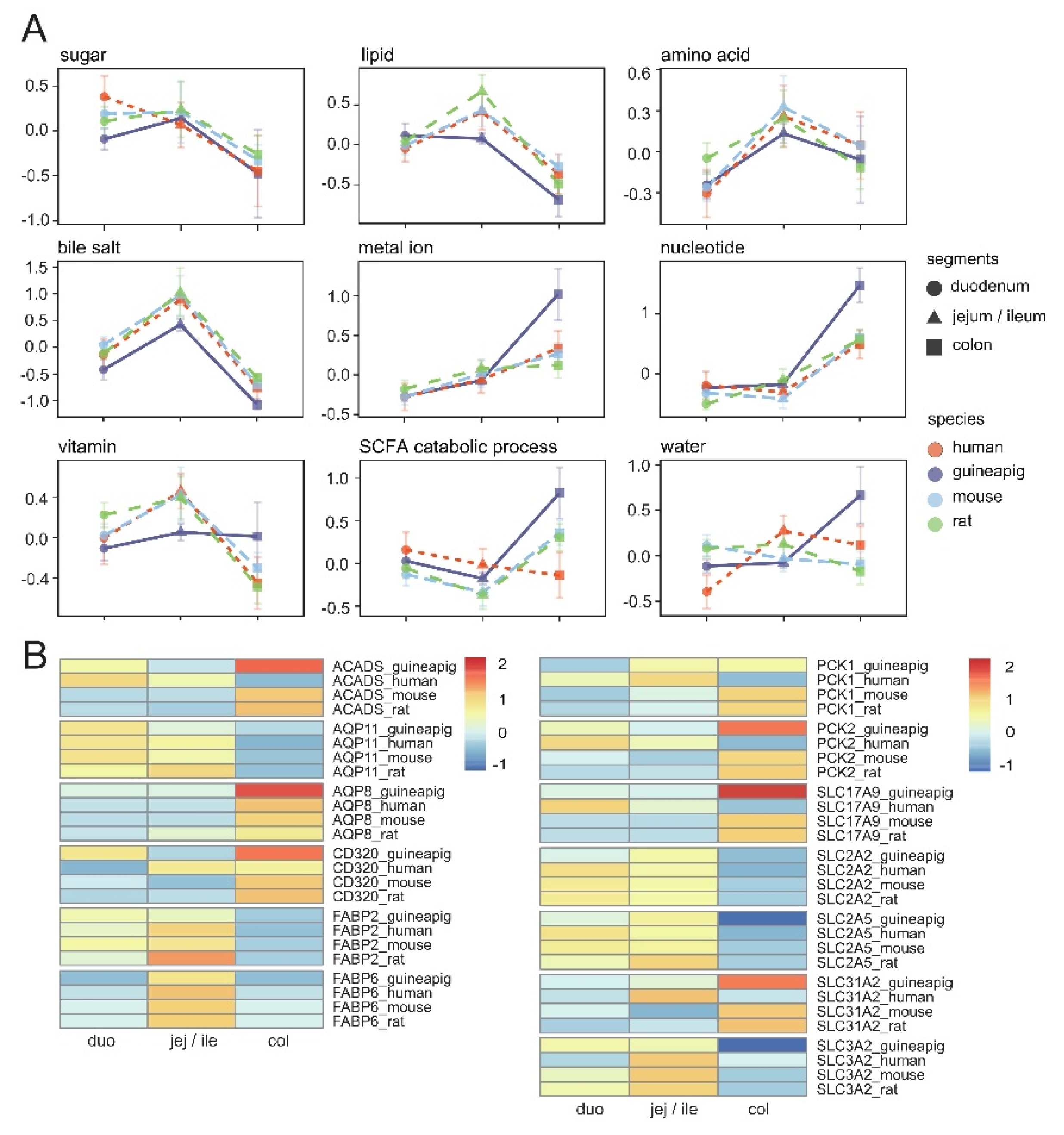
3.5. Differences in Gene Expression Related to Absorption among Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, H.; Esterházy, D. Intestinal immune compartmentalization: Implications of tissue specific determinants in health and disease. Mucosal Immunol 2021, 14, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Zorn, A.M.; Wells, J.M. Vertebrate Endoderm Development and Organ Formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 221–251. [Google Scholar] [CrossRef] [PubMed]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667. [Google Scholar] [CrossRef] [PubMed]
- Sancho, E.; Batlle, E.; Clevers, H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol 2004, 20, 695–723. [Google Scholar] [CrossRef]
- Wang, Y.; Song, W.; Wang, J.; Wang, T.; Xiong, X.; Qi, Z.; Fu, W.; Yang, X.; Chen, Y.G. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2019, 217, 20191130. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. Embo Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Diether, N.; Willing, B. Microbial Fermentation of Dietary Protein: An Important Factor in Diet–Microbe–Host Interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef]
- Mörbe, U.M.; Jørgensen, P.B.; Fenton, T.M.; von Burg, N.; Riis, L.B.; Spencer, J.; Agace, W.W. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021, 14, 793–802. [Google Scholar] [CrossRef]
- Ermund, A.; Schutte, A.; Johansson, M.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. AJP Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Pirr, S.; Viemann, D. Host factors of favorable intestinal microbial colonization. Front. Immunol. 2020, 11, 584288. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell. Host. Microbe. 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.D.; Selwyn, F.P.; Cui, J.Y.; Klaassen, C.D. RNA-Seq Profiling of Intestinal Expression of Xenobiotic Processing Genes in Germ-Free Mice. Drug Metab. Dispos. Biol. Fate Chem. 2017, 45, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Larsson, E.; Tremaroli, V.; Lee, Y.S.; Koren, O.; Nookaew, I.; Fricker, A.; Nielsen, J.; Ley, R.E.; Bäckhed, F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 2012, 61, 1124–1131. [Google Scholar] [CrossRef]
- Turner, P.V. The role of the gut microbiota on animal model reproducibility. Anim. Model. Exp. Med. 2018, 1, 109–115. [Google Scholar] [CrossRef]
- Hildebrand, F.; Ebersbach, T.; Nielsen, H.; Li, X.; Sonne, S.; Bertalan, M.; Dimitrov, P.; Madsen, L.; Qin, J.; Wang, J. A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens). Bmc Genom. 2012, 13, 514. [Google Scholar] [CrossRef]
- Elfers, K.; Armbrecht, Y.; Mazzuoli-Weber, G. Good to Know: Baseline Data on Feed Intake, Fecal Pellet Output and Intestinal Transit Time in Guinea Pig as a Frequently Used Model in Gastrointestinal Research. Animals 2021, 11, 1593. [Google Scholar] [CrossRef]
- Merchant, H.A.; Mcconnell, E.L.; Fang, L.; Ramaswamy, C.; Kulkarni, R.P.; Basit, A.W.; Murdan, S. Assessment of gastrointestinal pH, fluid and lymphoid tissue in the guinea pig, rabbit and pig, and implications for their use in drug development. Eur. J. Pharm. Sci. 2011, 42, 3–10. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, J.; Zhou, X.; Hu, S.; Li, M. Comprehensive Analysis of mRNA and lncRNA Transcriptomes Reveals the Differentially Hypoxic Response of Preadipocytes during Adipogenesis. Front Genet. 2020, 11, 845. [Google Scholar] [CrossRef]
- Feng, S.; Ma, J.; Long, K.; Zhang, J.; Qiu, W.; Li, Y.; Jin, L.; Wang, X.; Jiang, A.; Liu, L. Comparative microRNA transcriptomes in domestic goats reveal acclimatization to high altitude. Front. Genet. 2020, 809. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Söllner, J.F.; Leparc, G.; Hildebrandt, T.; Klein, H.; Thomas, L.; Stupka, E.; Simon, E. An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci. Data 2017, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Maertens, A.; Tran, V.; Kleensang, A.; Hartung, T. Weighted gene correlation network analysis (WGCNA) reveals novel transcription factors associated with bisphenol A dose-response. Front. Gene.t 2018, 9, 508. [Google Scholar] [CrossRef]
- Tappenden, K.A. Pathophysiology of short bowel syndrome: Considerations of resected and residual anatomy. J. Parenter. Enter. Nutr. 2014, 38, 14S–22S. [Google Scholar] [CrossRef]
- Sellge, G.; Kufer, T.A. PRR-signaling pathways–Learning from microbial tactics. Semin. Immunol. 2015, 27, 75–84. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Borrello, K.; Lim, U.; Park, S.; Monroe, K.R.; Maskarinec, G.; Boushey, C.J.; Wilkens, L.R.; Randolph, T.W.; Le Marchand, L.; Hullar, M.A. Dietary Intake Mediates Ethnic Differences in Gut Microbial Composition. Nutrients 2022, 14, 660. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ajami, N.J.; El-Serag, H.B.; Hair, C.; Graham, D.Y.; White, D.L.; Chen, L.; Wang, Z.; Plew, S.; Kramer, J. Dietary quality and the colonic mucosa–associated gut microbiome in humans. Am. J. Clin. Nutr. 2019, 110, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Xue, F.; Nan, X.; Tang, Z.; Wang, K.; Beckers, Y.; Jiang, L.; Xiong, B. Illumina sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front Microbiol. 2017, 8, 1818. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Li, J.; Li, T.; Zhang, Y.; Liang, Y.; Mei, Y. Phellinus linteus polysaccharide extract improves insulin resistance by regulating gut microbiota composition. Faseb. J. 2020, 34, 1065–1078. [Google Scholar] [CrossRef]
- Guo, M.; Li, Z. Polysaccharides isolated from Nostoc commune Vaucher inhibit colitis-associated colon tumorigenesis in mice and modulate gut microbiota. Food Funct. 2019, 10, 6873–6881. [Google Scholar] [CrossRef]
- Cao, X.; Gibbs, S.T.; Fang, L.; Miller, H.A.; Landowski, C.P.; Shin, H.; Lennernas, H.; Zhong, Y.; Amidon, G.L.; Yu, L.X. Why is it challenging to predict intestinal drug absorption and oral bioavailability in human using rat model. Pharm Res-Dordr. 2006, 23, 1675–1686. [Google Scholar] [CrossRef]
- Wang, J.; Cui, Y.; Xiao, L.; Chung, H.K.; Zhang, Y.; Rao, J.N.; Gorospe, M.; Wang, J. Regulation of intestinal epithelial barrier function by long noncoding RNA uc. 173 through interaction with microRNA 29b. Mol. Cell. Biol. 2018, 38, e10–e18. [Google Scholar]
- Qiao, Y.Q.; Huang, M.L.; Xu, A.T.; Zhao, D.; Ran, Z.H.; Shen, J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J. Biomed. Sci. 2013, 20, 1–7. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Pan, X.; Liu, M.; Wang, S.; Huang, T.; Cai, Y. Tissue expression difference between mRNAs and lncRNAs. Int. J. Mol. Sci. 2018, 19, 3416. [Google Scholar] [CrossRef]
- Romero, A.; Gomez, O.; Terrado, J.; Mesonero, J.E. Expression of GLUT8 in mouse intestine: Identification of alternative spliced variants. J. Cell. Biochem. 2009, 106, 1068–1078. [Google Scholar] [CrossRef]
- Helander, H.F.; Fändriks, L. Surface area of the digestive tract–revisited. Scand. J. Gastroentero. 2014, 49, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, Á.; Madsen, K.; Spiller, R.; Van Meerveld, B.G.; Verne, G.N. Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol. Motil. 2012, 24, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Hara, H. Dietary fat and bile juice, but not obesity, are responsible for the increase in small intestinal permeability induced through the suppression of tight junction protein expression in LETO and OLETF rats. Nutr. Metab. 2010, 7, 1–17. [Google Scholar] [CrossRef]
- Salama, N.N.; Eddington, N.D.; Fasano, A. Tight junction modulation and its relationship to drug delivery. Adv. Drug Deliver. Rev. 2006, 58, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Barone, S.; Fussell, S.L.; Singh, A.K.; Lucas, F.; Xu, J.; Kim, C.; Wu, X.; Yu, Y.; Amlal, H.; Seidler, U. Slc2a5 (Glut5) is essential for the absorption of fructose in the intestine and generation of fructose-induced hypertension. J. Biol. Chem. 2009, 284, 5056–5066. [Google Scholar] [CrossRef]
- Wright, E.M. Glucose transport families SLC5 and SLC50. Mol. Aspects. Med. 2013, 34, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Kellett, G.L.; Brot-Laroche, E.; Mace, O.J.; Leturque, A. Sugar absorption in the intestine: The role of GLUT2. Annu. Rev. Nutr. 2008, 28, 35–54. [Google Scholar] [CrossRef]
- Segade, F.; Allred, D.C.; Bowden, D.W. Functional characterization of the promoter of the human glucose transporter 10 gene. Biochim. Et Biophys. Acta. Gene. Struct. Expr. 2005, 1730, 147–158. [Google Scholar] [CrossRef]
- Perez-Martinez, P.; Delgado-Lista, J.; Perez-Jimenez, F.; Lopez-Miranda, J. Update on genetics of postprandial lipemia. Atheroscler. Supp. 2010, 11, 39–43. [Google Scholar] [CrossRef]
- Sztalryd, C.; Kimmel, A.R. Perilipins: Lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie 2014, 96, 96–101. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell. Bio. 2020, 21, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Buechler, C.; Aslanidis, C. Role of lipids in pathophysiology, diagnosis and therapy of hepatocellular carcinoma. Biochim. Biophys. Acta. Mol. Cell. Biol. Lipids 2020, 1865, 158658. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pochini, L.; Galluccio, M.; Console, L.; Indiveri, C. Glutamine transporters as pharmacological targets: From function to drug design. Asian J. Pharm. Sci. 2020, 15, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Monné, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial carriers for aspartate, glutamate and other amino acids: A review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, H.; Ning, L.; Li, F. Rabbit SLC15A1, SLC7A1 and SLC1A1 genes are affected by site of digestion, stage of development and dietary protein content. Animal 2019, 13, 326–332. [Google Scholar] [CrossRef]
- Feichtinger, R.G.; Lang, R. Targeting L-lactate metabolism to overcome resistance to immune therapy of melanoma and other tumor entities. J. Oncol. 2019, 1–12. [Google Scholar] [CrossRef]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef]
- Da Silva, T.C.; Polli, J.E.; Swaan, P.W. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol. Aspects Med. 2013, 34, 252–269. [Google Scholar] [CrossRef]
- Praslickova, D.; Torchia, E.C.; Sugiyama, M.G.; Magrane, E.J.; Zwicker, B.L.; Kolodzieyski, L.; Agellon, L.B. The ileal lipid binding protein is required for efficient absorption and transport of bile acids in the distal portion of the murine small intestine. PLoS ONE 2012, 7, e50810. [Google Scholar] [CrossRef]
- Wada, Y.; Abe, T.; Fuse, N.; Tamai, M. A frequent 1085delC/insGAAG mutation in the RDH5 gene in Japanese patients with fundus albipunctatus. Invest. Ophth. Vis. Sci. 2000, 41, 1894–1897. [Google Scholar]
- Weber, W.; Daoud-El Baba, M.; Fussenegger, M. Synthetic ecosystems based on airborne inter-and intrakingdom communication. Proc. Natl. Acad. Sci. USA 2007, 104, 10435–10440. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.J.; Pangilinan, F.J.; Cheng, J.; Molloy, A.M.; Brody, L.C. Mice lacking the transcobalamin-vitamin B12 receptor, CD320, suffer from anemia and reproductive deficits when fed vitamin B12-deficient diet. Hum. Mol. Genet. 2018, 27, 3627–3640. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Chen, Z.; Jiang, Z. Expression, distribution and role of aquaporin water channels in human and animal stomach and intestines. Int. J. Mol. Sci. 2016, 17, 1399. [Google Scholar] [CrossRef] [PubMed]
- Calamita, G.; Mazzone, A.; Bizzoca, A.; Cavalier, A.; Cassano, G.; Thomas, D.; Svelto, M. Expression and immunolocalization of the aquaporin-8 water channel in rat gastrointestinal tract. Eur. J. Cell. Biol. 2001, 80, 711–719. [Google Scholar] [CrossRef]
- Mao, S.; Fan, R.; Gu, T.; Zhong, Q.; Gong, M.; Dong, C.; Hao, L.; Zhang, L. Hypermethylation of SCNN1A gene-body increases the risk of essential hypertension. Int. J. Clin. Exp. Pathol 2016, 9, 8047–8056. [Google Scholar]
- Anderson, K.J.; Cormier, R.T.; Scott, P.M. Role of ion channels in gastrointestinal cancer. World. J. Gastroentero. 2019, 25, 5732. [Google Scholar] [CrossRef]
- Jonchère, V.; Brionne, A.; Gautron, J.; Nys, Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 2012, 12, 1–17. [Google Scholar] [CrossRef]
- Nebert, D.W.; Liu, Z. SLC39A8 gene encoding a metal ion transporter: Discovery and bench to bedside. Hum. Genom. 2019, 13, 1–21. [Google Scholar] [CrossRef]
- Murakami, T. Absorption sites of orally administered drugs in the small intestine. Expert Opin. Drug Dis. 2017, 12, 1219–1232. [Google Scholar] [CrossRef]
- Zhan, K.; Yang, T.Y.; Chen, Y.; Jiang, M.C.; Zhao, G.Q. Propionate enhances the expression of key genes involved in the gluconeogenic pathway in bovine intestinal epithelial cells. J. Dairy. Sci. 2020, 103, 5514–5524. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Vemula, P.K.; Jala, V.R. Colonic crypts are natural gatekeepers of microbial metabolites to protect stem cells. Transl. Cancer Res. 2016, 5, S536–S539. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, E.I. Digestive strategies of small hindgut fermenters. Anim. Sci. J. 2003, 74, 327–337. [Google Scholar] [CrossRef]
- Ferraris, R.P. Dietary and developmental regulation of intestinal sugar transport. Biochem. J. 2001, 360, 265–276. [Google Scholar] [CrossRef]
- Bouillon, R.; Van Cromphaut, S.; Carmeliet, G. Intestinal calcium absorption: Molecular vitamin D mediated mechanisms. J. Cell. Biochem. 2003, 88, 332–339. [Google Scholar] [CrossRef]
- Moreira, L.; Zamboni, D.S. NOD1 and NOD2 signaling in infection and inflammation. Front. Immunol. 2012, 3, 328. [Google Scholar] [CrossRef]
- Wang, Y.; Devkota, S.; Musch, M.W.; Jabri, B.; Nagler, C.; Antonopoulos, D.A.; Chervonsky, A.; Chang, E.B. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE 2010, 5, e13607. [Google Scholar] [CrossRef]
- Clarke, T.B.; Davis, K.M.; Lysenko, E.S.; Zhou, A.Y.; Yu, Y.; Weiser, J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010, 16, 228–231. [Google Scholar] [CrossRef]
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334, 255–258. [Google Scholar] [CrossRef]
- Habtezion, A.; Nguyen, L.P.; Hadeiba, H.; Butcher, E.C. Leukocyte trafficking to the small intestine and colon. Gastroenterology 2016, 150, 340–354. [Google Scholar] [CrossRef]
- Wendland, M.; Czeloth, N.; Mach, N.; Malissen, B.; Kremmer, E.; Pabst, O.; Förster, R. CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc. Natl. Acad. Sci. USA 2007, 104, 6347–6352. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.; Whon, T.W.; Bae, J. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Tsinganou, E.; Gebbers, J. Human intestinal spirochetosis–A review. GMS Ger. Med. Sci. 2010, 8. [Google Scholar] [CrossRef]
- Alenghat, T.; Osborne, L.C.; Saenz, S.A.; Kobuley, D.; Ziegler, C.G.; Mullican, S.E.; Choi, I.; Grunberg, S.; Sinha, R.; Wynosky-Dolfi, M. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 2013, 504, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Ichida, Y.; Hosokawa, N.; Takemoto, R.; Koike, T.; Nakatogawa, T.; Hiranuma, M.; Arakawa, H.; Miura, Y.; Azabu, H.; Ohtomo, S. Significant species differences in intestinal phosphate absorption between dogs, rats, and monkeys. J. Nutr. Sci. Vitaminol. 2020, 66, 60–67. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.; Ma, J.; Kong, F.; Li, B.; Du, Q.; Zhang, Y.; Wang, H.; Tang, Q.; Hu, S.; Liu, L.; et al. The Analysis of Transcriptomes and Microorganisms Reveals Differences between the Intestinal Segments of Guinea Pigs. Animals 2022, 12, 2925. https://doi.org/10.3390/ani12212925
Tang C, Ma J, Kong F, Li B, Du Q, Zhang Y, Wang H, Tang Q, Hu S, Liu L, et al. The Analysis of Transcriptomes and Microorganisms Reveals Differences between the Intestinal Segments of Guinea Pigs. Animals. 2022; 12(21):2925. https://doi.org/10.3390/ani12212925
Chicago/Turabian StyleTang, Chuang, Jideng Ma, Fanli Kong, Bo Li, Qinjiao Du, Yali Zhang, Haoming Wang, Qianzi Tang, Silu Hu, Lingyan Liu, and et al. 2022. "The Analysis of Transcriptomes and Microorganisms Reveals Differences between the Intestinal Segments of Guinea Pigs" Animals 12, no. 21: 2925. https://doi.org/10.3390/ani12212925
APA StyleTang, C., Ma, J., Kong, F., Li, B., Du, Q., Zhang, Y., Wang, H., Tang, Q., Hu, S., Liu, L., Li, X., & Li, M. (2022). The Analysis of Transcriptomes and Microorganisms Reveals Differences between the Intestinal Segments of Guinea Pigs. Animals, 12(21), 2925. https://doi.org/10.3390/ani12212925




