Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
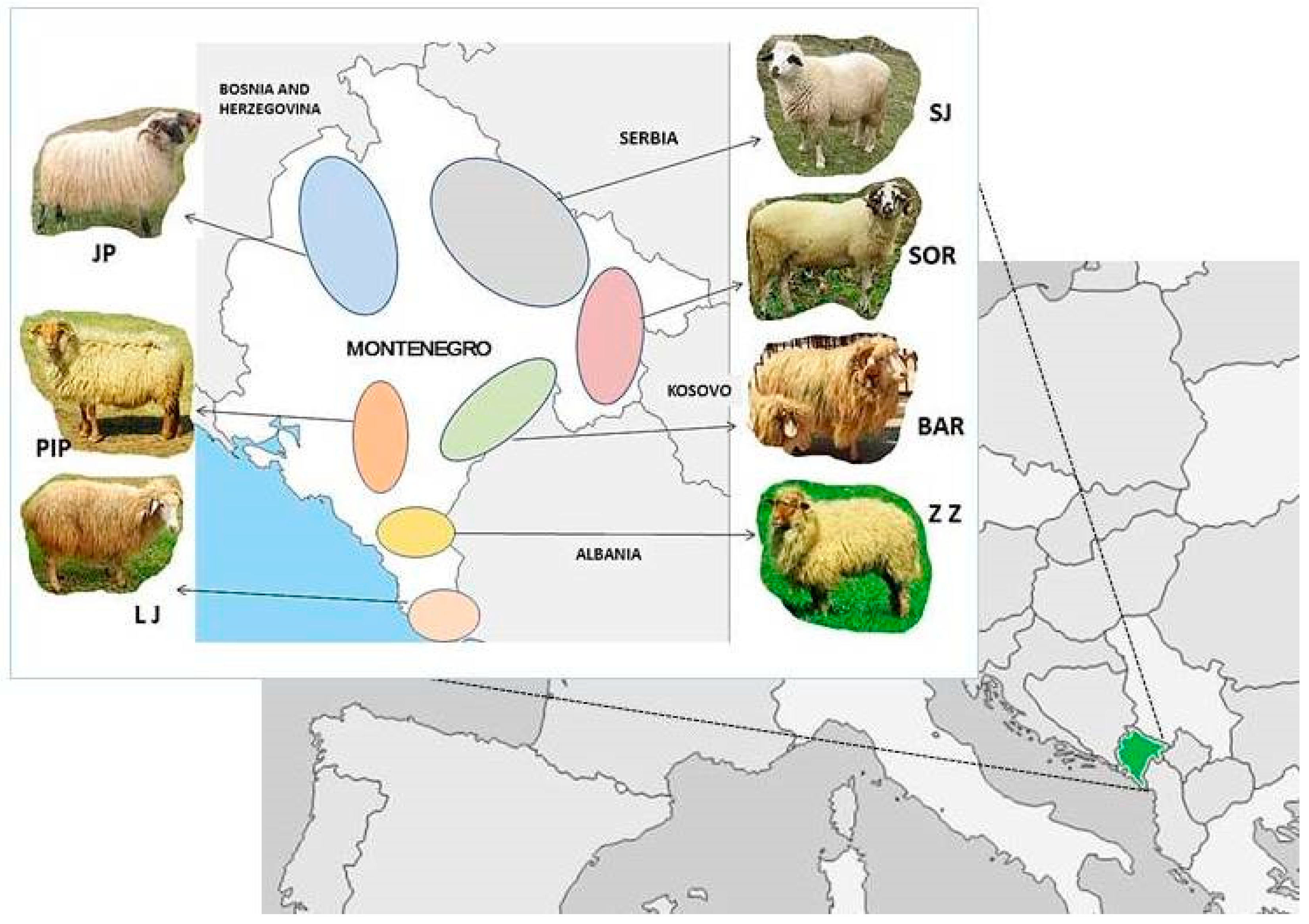
2.1. Animals
2.2. Sampling
2.3. Laboratory Procedures and Microsatellite Quality Control
2.4. Data Analyses
3. Results
3.1. Population Genetic Diversity of Microsatellite Locus
3.2. Genetic Diversity between Sheep Breeds
3.3. Genetic Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Scherf, B.D., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2015; Available online: http://www.fao.org/3/a-i4787e/index.html (accessed on 25 August 2022).
- Bowles, D. Recent advances in understanding the genetic resources of sheep breeds locally-adapted to the UK uplands: Opportunities they offer for sustainable productivity. Front. Genet. 2015, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Hristova, D.; Todorovska, E.; Vassilev, D.; Metodiev, S.; Popov, I.; Yablanski, T.; Zhelyazkov, E. Microsatellites based genetic diversity and population structure of seven Bulgarian indigenous sheep breeds. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 569–581. [Google Scholar]
- Juvančič, L.; Slabe-Erker, R.; Ogorevc, M.; Drucker, A.G.; Erjavec, E.; Bojkovski, D. Payments for Conservation of Animal Genetic Resources in Agriculture: One Size Fits All? Animals 2021, 11, 846. [Google Scholar] [CrossRef]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.M.; Toro, A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 41 (Suppl. S1), 6–31. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.A.; Meuwissen, T.H.E.; Fernandez, J.; Shaata, I.; Maki-Tanila, A. Assessing the genetic diversity in small farm animal populations. Animal 2011, 5, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Kusza, S.; Dimov, D.; Nagy, I.; Bõsze, S.; Jávor, A.; Kukovics, S. Microsatellite analysis to estimate genetic relationships among five Bulgarian sheep breeds. Genet. Mol. Biol. 2010, 33, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Baumung, R.; Cubric-Curik, V.; Schwend, K.; Achmann, R.; Solkner, J. Genetic characterization and breed assignment in Austrian sheep breeds using microsatellite marker information. J. Anim. Breed. Genet. 2006, 123, 265–271. [Google Scholar] [CrossRef]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311. [Google Scholar] [CrossRef]
- FAO; Rome. Molecular Genetic Characterization of Animal Genetic Resources. In FAO Animal Production and Health Guidelines; 2011; No. 9; Available online: http://www.fao.org/dad-is (accessed on 29 August 2022).
- Girish, H.; Sivaselvam, S.N.; Karthickeyan, S.M.K.; Saravanan, R. Molecular Characterisation of Nilagiri Sheep (Ovis aries) of South India Based on Microsatellites. Asian Aust. J. Anim. Sci. 2007, 20, 633–637. [Google Scholar] [CrossRef]
- Cinkulov, M.; Popovski, Z.; Tanaskovska, B.; Hodžić, A.; Bytyqi, H.; Mehmeti, H.; Margeta, V.; Djedović, R.; Hoda, A.; Trailović, R.; et al. Genetic diversity and structure of the West Balkan Pramenka sheep types as revealed by microsatellite and mitochondrial DNA analysis. J. Anim. Breed. Genet. 2008, 125, 417–426. [Google Scholar] [CrossRef]
- Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G.; Erhardt, G.; The Econogene Consortium. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim. Genet. 2007, 38, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Dudu, A.; Popa, G.O.; Ghit, E.; Pelmus, R.; Lazar, C.; Costache, M.; Georgescu, S.E. Assessment of genetic diversity in main local sheep breeds from Romania using microsatellite markers. Arch. Anim. Breed. 2020, 63, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, J.; Martinez, M.A.; Zaragoza, L.; Perezgrovas, R.; Vega-Pla, J.L.; Delgado, J.V. Genetic characterization of the autochthonous sheep populations from Chiapas, México. Livest. Sci. 2008, 116, 156–161. [Google Scholar] [CrossRef]
- Marina, H.; Suarez-Vega, A.; Pelayo, R.; Gutiérrez-Gil, B.; Reverter, A.; Esteban-Blanco, C.; Arranz, J.J. Accuracy of Imputation of Microsatellite Markers from a 50K SNP Chip in Spanish Assaf Sheep. Animals 2021, 11, 86. [Google Scholar] [CrossRef]
- Dossybayev, K.; Orazymbetova, Z.; Mussayeva, A.; Saitou, N.; Zhapbasov, R.; Makhatov, B.; Bekmanov, B. Genetic diversity of different breeds of Kazakh sheep using microsatellite analysis. Arch. Anim. Breed. 2019, 62, 305–312. [Google Scholar] [CrossRef]
- Abdelkader, A.A.; Ata, N.; Benyoucef, M.T.; Djaout, A.; Azzi, N.; Yilmaz, O.; Cemal, I.; Gaouar, S.B. New genetic identification and characterisation of 12 Algerian sheep breeds by microsatellite markers. Ital. J. Anim. Sci. 2018, 17, 38–48. [Google Scholar] [CrossRef]
- Salinas-Rios, T.; Hernández-Bautista, J.; Mariscal-Méndez, A.; Aquino-Cleto, M.; Martínez-Martínez, A.; Rodríguez-Magadán, H.M. Genetic Characterization of a Sheep Population in Oaxaca, Mexico: The Chocholteca Creole. Animals 2021, 11, 1172. [Google Scholar] [CrossRef]
- MONSTAT. Statistical Yearbook of Montenegro; Statistical Office of Montenegro SY MNE. , 2021; pp. 110–111. Available online: www.monstat.org (accessed on 27 October 2022).
- Porcu, K.; Marković, B. Catalogue of West Balkan Pramenka Sheep Breed Type; Zemljodelski fakultet: Skopje, Macedeonia, 2006; p. 90. ISBN 9989-845-23-9. [Google Scholar]
- Marković, B.; Dovč, P.; Marković, M.; Radonjić, D.; Adakalić, M.; Simčič, M. Differentiation of some Pramenka sheep breeds based on morphometric characteristics. Arch. Anim. Breed 2019, 62, 393–402. [Google Scholar] [CrossRef]
- Marković, B.; Marković, M.; Adžić, N. Genetic Resources in Livestock of Montenegro; Montenegrin Academy of Science and Art: Podgorica, Montenegro, 2020; p. 300. ISBN 978-86-7215-470-2. [Google Scholar]
- Ivanković, A.; Dovč, P. Polimorfizem genov za β-laktoglobulin in αs1-kazein paske ovce. Acta Agric. Slov. 2004, 842, 121–130. [Google Scholar]
- ISAG Conference Dublin, Ireland. Recommended Sheep and Goats Panels of Microsatellite Markers. 2017. Available online: https://www.isag.us/Docs/AppGenSheepGoat2017.pdf (accessed on 17 July 2022).
- Currie-Fraser, E.; Shah, P.; True, S. Data analysis using GeneMapper® v4.1: Comparing the Newest Generation of GeneMapper Software to Legacy Genescan® and Genotyper® Software. J. Biomol. Tech. 2010, 21 (Suppl. S3), S31. [Google Scholar]
- Valiere, N. GIMLET: A computer program for analyzing genetic individual identification data. Mol. Ecol. Notes 2002, 2, 377–379. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Bio. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C.; et al. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. 2020, 52, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Đokić, M.; Marković, B.; Gantner, V.; Marković, M. Association of genetic variants of β-lactoglobulin gene with milk traits of Jezeropivska sheep breed. Agric. For. 2020, 66, 15–23. [Google Scholar] [CrossRef]
- Mihailova, Y. Genetic diversity and structure of 2 indigenous sheep breeds (Kotel and Teteven) in Bulgaria using microsatellite markers. Biotechnol. Biotechnol. Equip. 2021, 35, 576–585. [Google Scholar] [CrossRef]
- Karsli, B.A.; Demir, E.; Fidan, H.G.; Karsli, T. Genetic diversity and differentiation among Turkish sheep breeds. Arch. Anim. Breed. 2020, 63, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Ćurković, M.; Ramljak, J.; Ivanković, S.; Mioc, B.; Ivankovic, A.; Pavic, V.; Brka, M.; Veit-Kensch, C.; Medugorac, I. The genetic diversity and structure of 18 sheep breeds exposed to isolation and selection. J. Anim. Breed. Genet. 2016, 133, 71–80. [Google Scholar] [CrossRef]
- Sharma, R.; Ahlawat, S.; Sharma, H.; Sharma, P.; Panchal, P.; Arora, R.; Tantia, M.S. Microsatellite and mitochondrial DNA analyses unveil the genetic structure of native sheep breeds from three major agro-ecological regions of India. Sci. Rep. 2020, 10, 20422. [Google Scholar] [CrossRef]
- Jawasreh, K.I.; Ababneh, M.M.; Ismail, Z.B.; Younes, A.M.B.; Al Sukhni, I. Genetic diversity and population structure of local and exotic sheep breeds in Jordan using microsatellites markers. Vet. World 2018, 11, 778–781. [Google Scholar] [CrossRef]
- Salamon, D.; Gutierrez-Gil, B.; Arranz, J.J.; Barreta, J.; Batinic, V.; Dzidic, A. Genetic diversity and differentiation of 12 eastern Adriatic and western Dinaric native sheep breeds using microsatellites. Animal 2014, 8, 200–207. [Google Scholar] [CrossRef]
- Al-Atiyat, R.M.; Aljumaah, R.S.; Alshaikh, M.A.; Abudabos, A.M. Microsatellite-Based Genetic Structure and Diversity of Local Arabian Sheep Breeds. Front. Genet. 2018, 9, 408. [Google Scholar] [CrossRef]
- Vajed Ebrahimi, M.T.; Mohammadabadi, M.; Esmailizadeh, A. Using microsatellite markers to analyze genetic diversity in 14 sheep types in Iran. Arch. Anim. Breed. 2017, 60, 183–189. [Google Scholar] [CrossRef]
- Carlsson, J. Effects of Microsatellite Null Alleles on Assignment Testing. J. Hered. 2008, 99, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Oner, J.; Ustuner, H.; Orman, A.; Yilmaz, O.; Yılmaz, A. Genetic Diversity of Kıvırcık Sheep Breed Reared in Different Regions and its Relationship with Other Sheep Breeds in Turkey. Ital. J. Anim. Sci. 2014, 13, 3382. [Google Scholar] [CrossRef]



| Name of Breed | Acronym of Breed | WH | BL | CD | CW | CC | CBC | BW |
|---|---|---|---|---|---|---|---|---|
| Jezeropivska | JP | 71.3 | 70.8 | 32.1 | 21.7 | 110.1 | 9.0 | 71.6 |
| Sora | SOR | 69.7 | 69.9 | 30.6 | 20.3 | 99.2 | 9.1 | 63.8 |
| Zetaska zuja | ZZ | 63.1 | 59.4 | 27.3 | 15.1 | 81.8 | 7.6 | 37.1 |
| Bardoka | BAR | 66.2 | 66.2 | 29.1 | 18.3 | 92.1 | 8.4 | 54.3 |
| Ljaba | LJ | 63.4 | 62.2 | 29.0 | 18.8 | 87.4 | 8.1 | 46.5 |
| Sjenička | SJ | 72.7 | 77.9 | 33.1 | 21.3 | 100.3 | 8.9 | 77.3 |
| Piperska zuja | PIP | 68.8 | 65.4 | 31.4 | 21.2 | 96.8 | 8.4 | 52.7 |
| Marker | Chr. | Primer (5′–3′) | Annealing, °C | Size (bp) |
|---|---|---|---|---|
| OarAE 129 | 5 | F: AATCCAGTGTGTGAAAGACTAATCCAG R: GTAGATCAAGATATAGAATATTTTTCAACACC | 62 | 135–161 |
| OarCP49 | 17 | F: CAGACACGGCTTAGCAACTAAACGC R: GTGGGGATGAATATTCCTTCATAAGG | 64 | 80–112 |
| CSRD247 | 14 | F: GGACTTGCCAGAACTCTGCAAT R:CACTGTGGTTTGTATTAGTCAGG | 58 | 220–246 |
| OarFCB20 | 2 | F: GGAAAACCCCCATATATACCTATAC R: AATGTGTTTAAGATTCCATACATGTG | 60 | 87–115 |
| HSC | 20 | F: CTGCCAATGCAGAGACACAAGA R: GTCTGTCTCCTGTCTTGTCATC | 63 | 263–297 |
| ILST05 | 7 | F: GGAAGCAATGAAATCTATAGCC R: TGTTCTGTGAGTTTGTAAGC | 55 | 194–254 |
| ILSTS011 | 9 | F: GCTTGCTACATGGAAAGTGC R: CTAAAATGCAGAGCCCTACC | 55 | 250–300 |
| INRA006 | 1 | F: AGGAATATCTGTATCAACCGCAGTC R: CTGAGCTGGGGTGGGAGCTATAAATA | 64 | 110–132 |
| INRA023 | 3 | F: GAGTAGAGCTACAAGATAAACTTC R: TAACTACAGGGTGTTAGATGAACTC | 58 | 194–216 |
| INRA063 | 14 | F: GACCACAAAGGGATTTGCACAAGC R: AAACCACAGAAATGCTTGGAAG | 56 | 169–201 |
| INRA049 | 1 | F: TGTATTAGTTTGTGTTCTTTGGC R: TTGGCTTCCACAATCACACA | 61 | 134–166 |
| INRA132 | 20 | F: AACATTTCAGCTGATGGTGGC R: TTCTGTTTTGAGTGGTAAGCT G | 62 | 146–180 |
| INRA172 | 22 | F: CCAGGGCAGTAAAATGCATAACTG R: GGCCTTGCTAGCCTCTGCAAAC | 65 | 126–172 |
| MAF065 | 15 | F: AAAGGCCAGAGTATGCAATTAGGAG R: CCACTCCTCCTGAGAATATAACATG | 59 | 116–158 |
| MAF214 | 16 | F: AATGCAGGAGATCTGAGGCAGGGACG R: GGGTGATCTTAGGGAGGTTTTGGAGG | 66 | 189–265 |
| McM042 | 9 | F: GTTCGTACTTCTGGGTACTGGTCTC R: GTCCATGGATTTGCAGAGTCAG | 60 | 81–107 |
| SPS113 | 10 | F: CCTCCACACAGGCTTCTCTGACTT R: CCTAACTTGCTTGAGTTATTGCCC | 60 | 126–152 |
| SPS115 | 15 | F: AAAGTGACACAACAGCTTCTCCAG R: AACGAGTGTCCTAGTTTGGCTGTG | 62 | 246–260 |
| Locus | Na | No | Ho | He | uHe | HWD | F | PIC |
|---|---|---|---|---|---|---|---|---|
| AE129a | 6 | 0.034 | 0.626 | 0.642 | 0.650 | 0.481 ns | 0.013 | 0.676 |
| CP49a | 25 | 0.026 | 0.816 | 0.794 | 0.804 | 0.942 ns | −0.031 | 0.938 |
| CSRD247a | 17 | 0.017 | 0.863 | 0.825 | 0.836 | 0.003 ** | −0.047 | 0.866 |
| FCB20a | 14 | 0.033 | 0.780 | 0.770 | 0.779 | 0.016 ns | −0.014 | 0.918 |
| HSCa | 16 | 0.027 | 0.812 | 0.815 | 0.826 | 0.012 ns | 0.002 | 0.868 |
| ILSTS005a | 11 | 0.050 | 0.672 | 0.693 | 0.702 | 0.000 ** | 0.032 | 0.712 |
| ILSTS011a | 8 | 0.039 | 0.687 | 0.688 | 0.697 | 0.276 ns | −0.012 | 0.724 |
| INRA006a | 11 | 0.016 | 0.723 | 0.685 | 0.694 | 0.196 ns | −0.059 | 0.720 |
| INRA023a | 13 | 0.046 | 0.799 | 0.794 | 0.804 | 0.000 ** | −0.010 | 0.876 |
| INRA049a | 8 | 0.040 | 0.673 | 0.677 | 0.686 | 0.000 ** | 0.012 | 0.704 |
| INRA063a | 21 | 0.039 | 0.744 | 0.758 | 0.768 | 0.000 ** | 0.027 | 0.794 |
| INRA132a | 14 | 0.024 | 0.833 | 0.815 | 0.826 | 0.025 ns | −0.021 | 0.866 |
| INRA172a | 17 | 0.041 | 0.573 | 0.590 | 0.598 | 0.010 ns | 0.032 | 0.602 |
| MAF214a | 14 | 0.023 | 0.681 | 0.701 | 0.710 | 0.404 ns | 0.027 | 0.714 |
| MAF65a | 11 | 0.020 | 0.756 | 0.741 | 0.750 | 0.240 ns | −0.026 | 0.770 |
| McM042a | 11 | 0.019 | 0.662 | 0.627 | 0.635 | 0.983 ns | −0.062 | 0.843 |
| SPS113a | 12 | 0.000 | 0.680 | 0.638 | 0.646 | 0.002 ** | −0.069 | 0.653 |
| SPS115a | 14 | 0.056 | 0.732 | 0.767 | 0.777 | 0.003 ** | 0.043 | 0.821 |
| MEAN | 13.5 | 0.031 | 0.728 | 0723 | 0.733 | 0.199 | −0.009 | 0.781 |
| Locus | Fis | Fit | Fst | Gst | Ht | D |
|---|---|---|---|---|---|---|
| AE129a | 0.024 | 0.121 | 0.099 | 0.088 | 0.712 | 0.211 |
| CP49a | −0.029 | 0.080 | 0.105 | 0.095 | 0.887 | 0.503 |
| CSRD247a | −0.046 | 0.016 | 0.059 | 0.049 | 0.877 | 0.305 |
| FCB20a | −0.013 | 0.095 | 0.106 | 0.096 | 0.861 | 0.439 |
| HSCa | 0.004 | 0.072 | 0.068 | 0.057 | 0.875 | 0.338 |
| ILSTS005a | 0.031 | 0.106 | 0.078 | 0.067 | 0.752 | 0.197 |
| ILSTS011a | 0.001 | 0.104 | 0.103 | 0.092 | 0.767 | 0.273 |
| INRA006a | −0.055 | 0.027 | 0.078 | 0.068 | 0.743 | 0.193 |
| INRA023a | −0.006 | 0.094 | 0.100 | 0.089 | 0.882 | 0.471 |
| INRA049a | 0.005 | 0.111 | 0.106 | 0.096 | 0.757 | 0.269 |
| INRA063a | 0.018 | 0.094 | 0.078 | 0.067 | 0.822 | 0.277 |
| INRA132a | −0.022 | 0.051 | 0.071 | 0.060 | 0.877 | 0.355 |
| INRA172a | 0.029 | 0.091 | 0.063 | 0.052 | 0.630 | 0.096 |
| MAF214a | 0.028 | 0.093 | 0.066 | 0.055 | 0.751 | 0.167 |
| MAF65a | −0.021 | 0.054 | 0.074 | 0.063 | 0.800 | 0.236 |
| McM042a | −0.057 | 0.022 | 0.075 | 0.065 | 0.677 | 0.140 |
| SPS113a | −0.065 | 0.013 | 0.074 | 0.064 | 0.689 | 0.145 |
| SPS115a | 0.046 | 0.128 | 0.085 | 0.074 | 0.839 | 0.326 |
| MEAN | −0.007 | 0.076 | 0.083 | 0.072 | 0.789 | 0.250 |
| Breed | Acronym | N | Na | NE | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|---|
| Jezeropivska | JP | 60 | 10.056 | 4.972 | 1.772 | 0.763 | 0.772 | 0.778 | 0.016 |
| Sora | SOR | 44 | 9.500 | 4.705 | 1.732 | 0.754 | 0.762 | 0.771 | 0.011 |
| Zeta zuja | ZZ | 33 | 7.722 | 3.708 | 1.515 | 0.747 | 0.704 | 0.715 | −0.064 |
| Bardoka | BAR | 43 | 8.667 | 4.603 | 1.658 | 0.690 | 0.743 | 0.752 | 0.074 |
| Ljaba | LJ | 44 | 8.278 | 4.248 | 1.608 | 0.740 | 0.732 | 0.740 | −0.015 |
| Sjenička | SJ | 44 | 7.889 | 3.703 | 1.522 | 0.717 | 0.711 | 0.719 | −0.007 |
| Piperska zuja | PIP | 23 | 4.889 | 3.195 | 1.260 | 0.688 | 0.640 | 0.654 | −0.079 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marković, M.; Radonjić, D.; Zorc, M.; Đokić, M.; Marković, B. Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers. Animals 2022, 12, 3029. https://doi.org/10.3390/ani12213029
Marković M, Radonjić D, Zorc M, Đokić M, Marković B. Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers. Animals. 2022; 12(21):3029. https://doi.org/10.3390/ani12213029
Chicago/Turabian StyleMarković, Milan, Dušica Radonjić, Minja Zorc, Milena Đokić, and Božidarka Marković. 2022. "Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers" Animals 12, no. 21: 3029. https://doi.org/10.3390/ani12213029
APA StyleMarković, M., Radonjić, D., Zorc, M., Đokić, M., & Marković, B. (2022). Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers. Animals, 12(21), 3029. https://doi.org/10.3390/ani12213029





