Yeast Products Mediated Ruminal Subenvironmental Microbiota, and Abnormal Metabolites and Digestive Enzymes Regulated Rumen Fermentation Function in Sheep
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals and Experimental Design
2.3. Sample Collection and Processing
2.4. Digestive Enzyme Activities and Abnormal Metabolites
2.5. Fermentation Parameters
2.6. Microbial DNA Extraction and Metagenomic Sequencing
2.7. Pyrosequencing Data Accession Number
2.8. Statistical Analysis
3. Results
3.1. Effects of YP on Fermentation Parameters, Abnormal Metabolites and Digestive Enzyme Activity
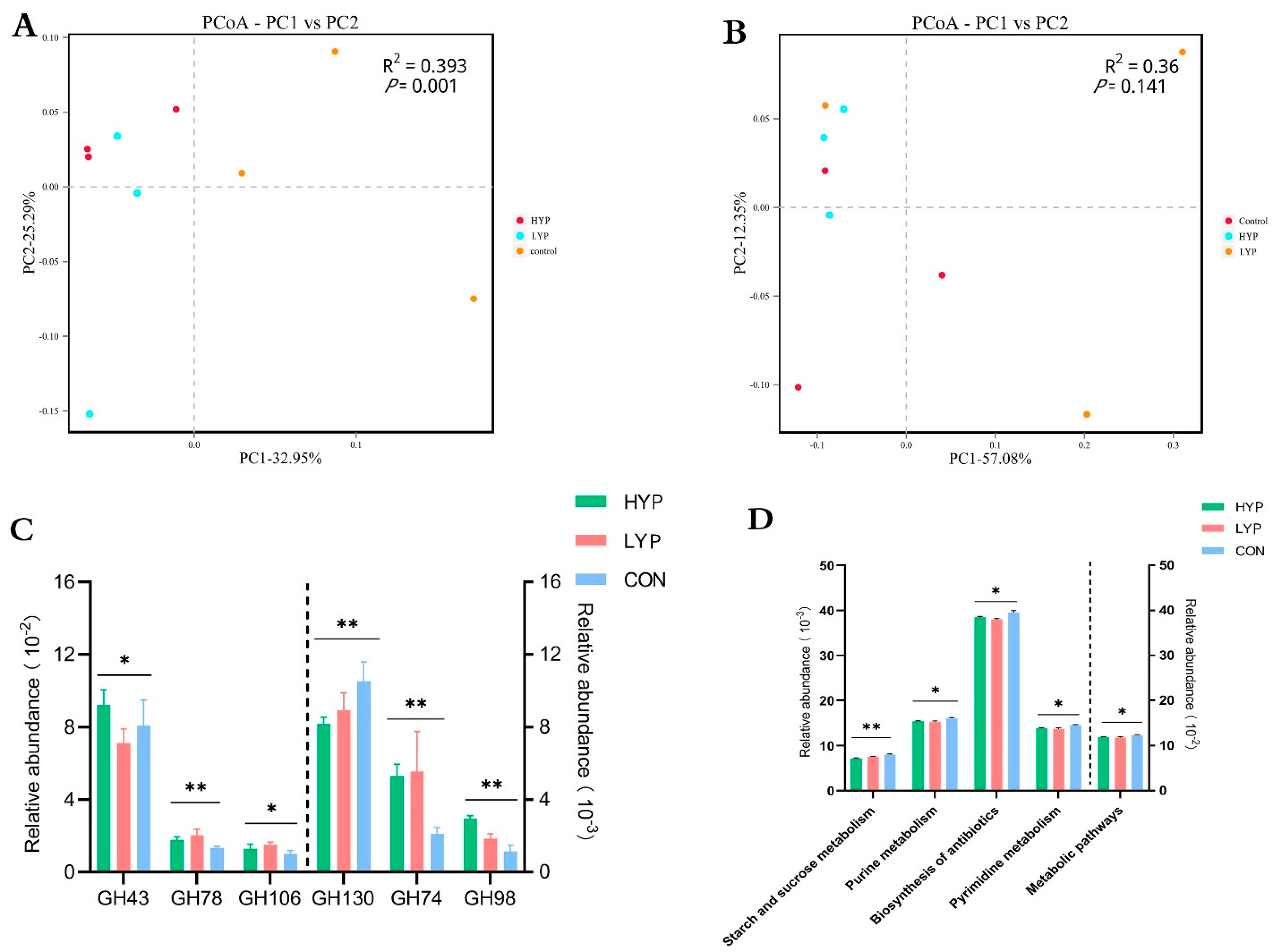
3.2. YP Changed the Composition Profiles of Rumen Solid-Associated (SA) and Liquid-Associated (LA) Microbiota
3.3. YP Changed the Composition of Microorganisms Related to Lactic Acid Metabolism and Methanogens in the Rumen
3.4. YP Changed Functional Profiles of Rumen Solid-Associated (SA) and Liquid-Associated (LA) Microbiota
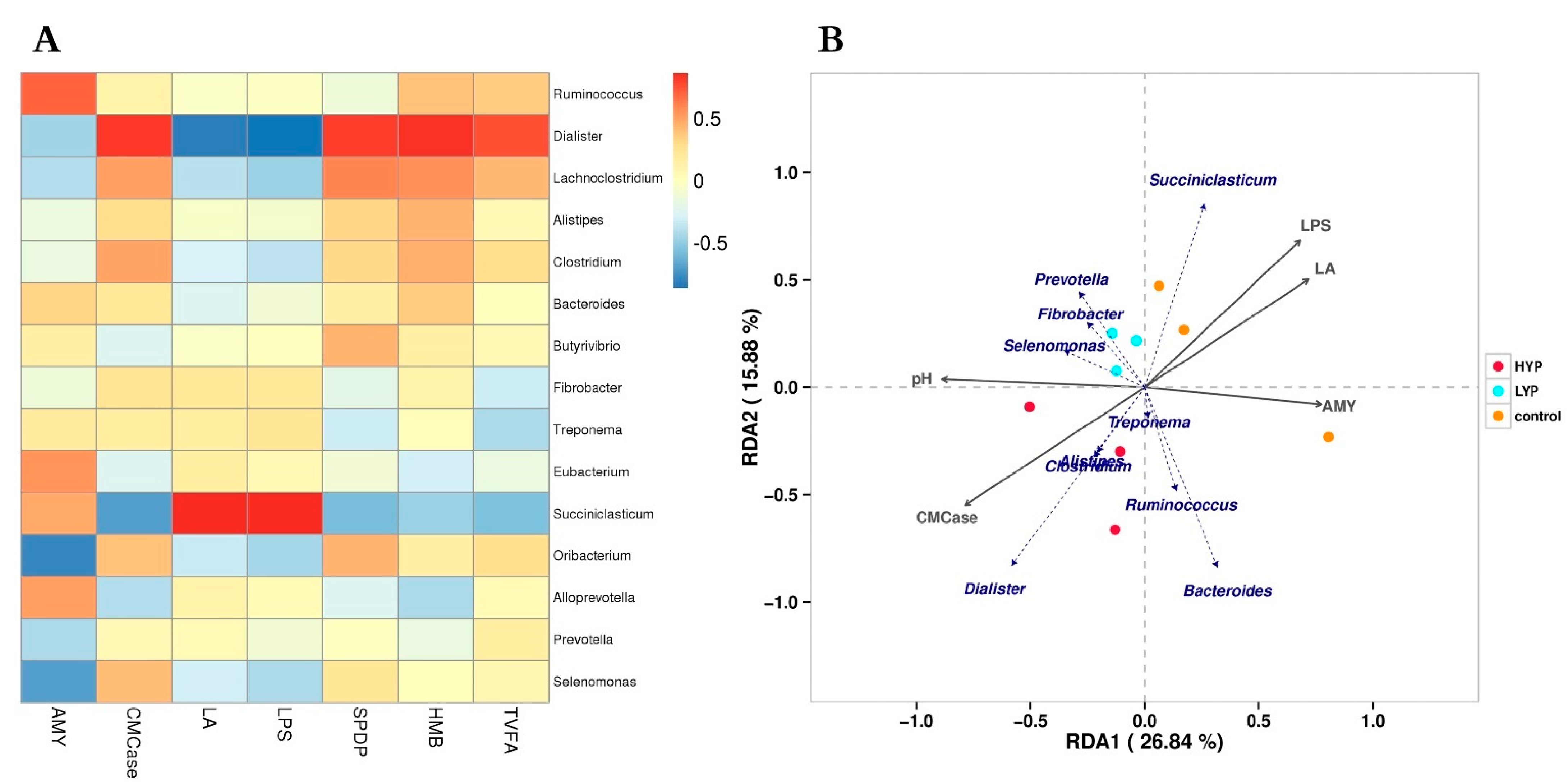
3.5. Correlation Analysis between Rumen Digestive Enzyme, Fermentation Parameters, Abnormal Metabolites and Microbiota
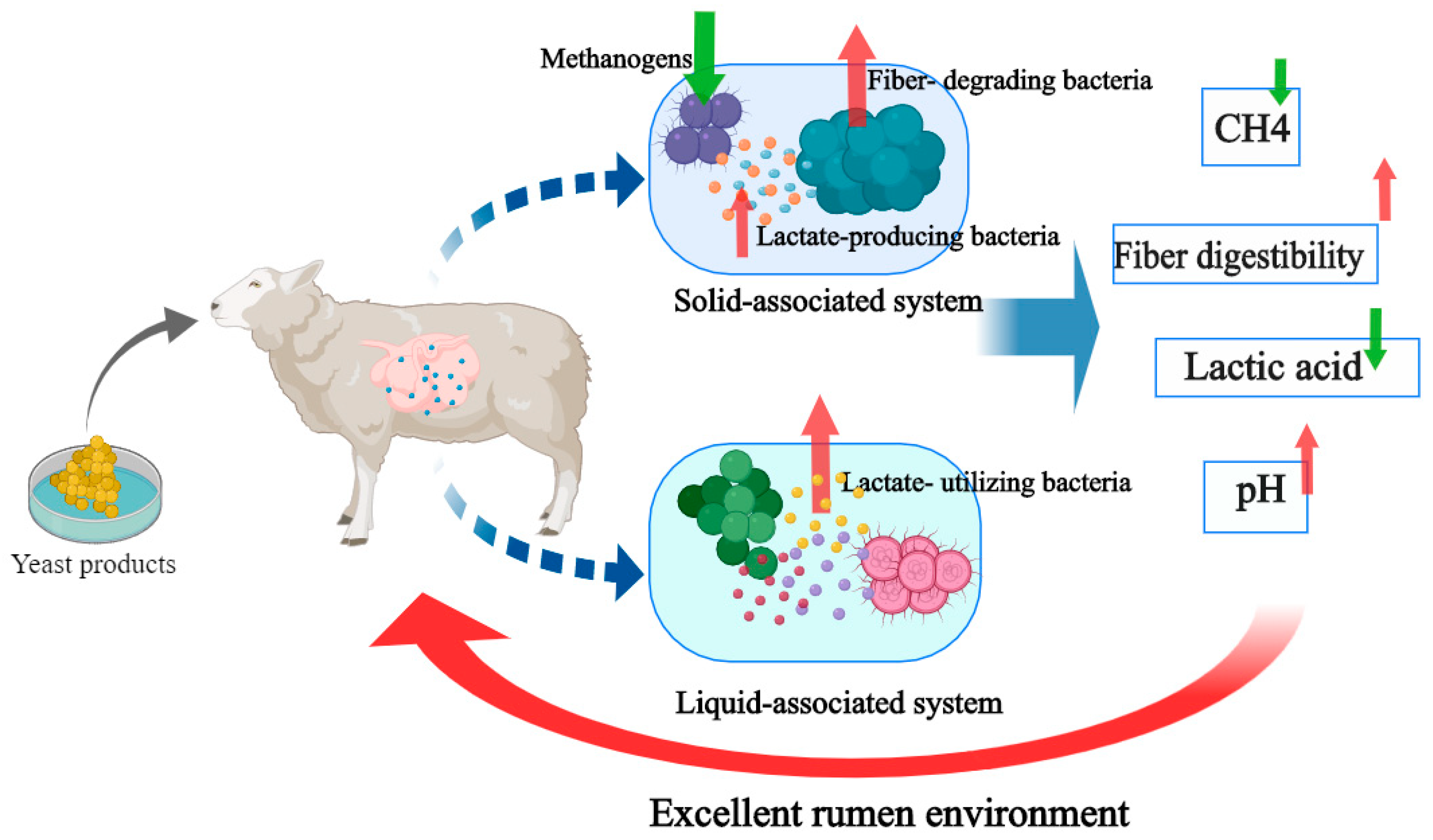
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silberberg, M.; Chaucheyras-Durand, F.; Commun, L.; Mialon, M.; Monteils, V.; Mosoni, P.; Morgavi, D.; Martin, C. Repeated acidosis challenges and live yeast supplementation shape rumen microbiota and fermentations and modulate inflammatory status in sheep. Animal 2013, 7, 1910–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAllister, T.A.; Beauchemin, K.A.; Alazzeh, A.Y.; Baah, J.; Teather, R.M.; Stanford, K. Review: The use of direct fed microbials to mitigate pathogens and enhance production in cattle. Can. J. Anim. Sci. 2011, 91, 193–211. [Google Scholar] [CrossRef] [Green Version]
- Elghandour, M.M.; Khusro, A.; Adegbeye, M.J.; Tan, Z.; Abu Hafsa, S.H.; Greiner, R.; Ugbogu, E.A.; Anele, U.Y.; Salem, A.Z.M. Dynamic role of single-celled fungi in ruminal microbial ecology and activities. J. Appl. Microbiol. 2020, 128, 950–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.; Luo, J.; Rayward, D.; King, S.; Gibson, R.; Moghaddam, G. Selection of a novel direct-fed microbial to enhance weight gain in intensively reared calves. Anim. Feed Sci. Technol. 2008, 145, 41–52. [Google Scholar] [CrossRef]
- Geng, C.-Y.; Ren, L.-P.; Zhou, Z.-M.; Chang, Y.; Meng, Q.-X. Comparison of active dry yeast (Saccharomyces cerevisiae) and yeast culture for growth performance, carcass traits, meat quality and blood indexes in finishing bulls. Anim. Sci. J. 2015, 87, 982–988. [Google Scholar] [CrossRef]
- Sales, J. Effects of Saccharomyces cerevisiae supplementation on ruminal parameters, nutrient digestibility and growth in sheep: A meta-analysis. Small Rumin. Res. 2011, 100, 19–29. [Google Scholar] [CrossRef]
- Bayat, A.; Kairenius, P.; Stefański, T.; Leskinen, H.; Comtet-Marre, S.; Forano, E.; Chaucheyras-Durand, F.; Shingfield, K. Effect of camelina oil or live yeasts (Saccharomyces cerevisiae) on ruminal methane production, rumen fermentation, and milk fatty acid composition in lactating cows fed grass silage diets. J. Dairy Sci. 2015, 98, 3166–3181. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Ogunade, I.M.; Qi, S.; Hackmann, T.J.; Staples, C.R.; Adesogan, A.T. Effects of the dose and viability of Saccharomyces cerevisiae. 1. Diversity of ruminal microbes as analyzed by Illumina MiSeq sequencing and quantitative PCR. J. Dairy Sci. 2017, 100, 325–342. [Google Scholar] [CrossRef] [Green Version]
- Amin, A.B.; Mao, S. Influence of yeast on rumen fermentation, growth performance and quality of products in ruminants: A review. Anim. Nutr. 2020, 7, 31–41. [Google Scholar] [CrossRef]
- Pinloche, E.; McEwan, N.; Marden, J.-P.; Bayourthe, C.; Auclair, E.; Newbold, C.J. The Effects of a Probiotic Yeast on the Bacterial Diversity and Population Structure in the Rumen of Cattle. PLoS ONE 2013, 8, e67824. [Google Scholar] [CrossRef]
- Mao, H.-L.; Wang, J.K.; Liu, J.X.; Yoon, I. Effects of Saccharomyces cerevisiae fermentation product on in vitro fermentation and microbial communities of low-quality forages and mixed diets. J. Anim. Sci. 2013, 91, 3291–3298. [Google Scholar] [CrossRef] [PubMed]
- Ogunade, I.; Schweickart, H.; McCoun, M.; Cannon, K.; McManus, C. Integrating 16S rRNA Sequencing and LC–MS-Based Metabolomics to Evaluate the Effects of Live Yeast on Rumen Function in Beef Cattle. Animals 2019, 9, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darabighane, B.; Salem, A.Z.M.; Aghjehgheshlagh, F.M.; Mahdavi, A.; Zarei, A.; Elghandour, M.M.M.Y.; López, S. Environmental efficiency of Saccharomyces cerevisiae on methane production in dairy and beef cattle via a meta-analysis. Environ. Sci. Pollut. Res. 2018, 26, 3651–3658. [Google Scholar] [CrossRef]
- Lu, Q.; Wu, J.; Wang, M.; Zhou, C.; Han, X.; Odongo, E.N.; Tan, Z.; Tang, S. Effects of dietary addition of cellulase and a Saccharomyces cerevisiae fermentation product on nutrient digestibility, rumen fermentation and enteric methane emissions in growing goats. Arch. Anim. Nutr. 2016, 70, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Hao, Z.; Shi, H.; Li, T.; Wang, H.; Li, Q.; Zhang, Y.; Ma, Y. Metagenomic Analysis Revealed Differences in Composition and Function Between Liquid-Associated and Solid-Associated Microorganisms of Sheep Rumen. Front. Microbiol. 2022, 13, 851567. [Google Scholar] [CrossRef]
- Broderick, G.A.; Kang, J.H. Automated Simultaneous Determination of Ammonia and Total Amino Acids in Ruminal Fluid and In Vitro Media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, J.; Lei, Y.; Bai, Y.; Jia, L.; Li, Z.; Liu, T.; Xu, Y.; Sun, J.; Wang, Y.; et al. Oregano Essential Oils Promote Rumen Digestive Ability by Modulating Epithelial Development and Microbiota Composition in Beef Cattle. Front. Nutr. 2021, 8, 722557. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Xiao, J.; Alugongo, G.; Chung, R.; Dong, S.; Li, S.; Yoon, I.; Wu, Z.; Cao, Z. Effects of Saccharomyces cerevisiae fermentation products on dairy calves: Ruminal fermentation, gastrointestinal morphology, and microbial community. J. Dairy Sci. 2016, 99, 5401–5412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Wei, Z.; Xu, N.; Yang, F.; Yoon, I.; Chung, Y.; Liu, J.; Wang, J. Effects of Saccharomyces cerevisiae fermentation products on performance and rumen fermentation and microbiota in dairy cows fed a diet containing low quality forage. J. Anim. Sci. Biotechnol. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Chaucheyras-Durand, F.; Walker, N.; Bach, A. Effects of active dry yeasts on the rumen microbial ecosystem: Past, present and future. Anim. Feed Sci. Technol. 2008, 145, 5–26. [Google Scholar] [CrossRef]
- Peng, Q.-H.; Cheng, L.; Kang, K.; Tian, G.; Al-Mamun, M.; Xue, B.; Wang, L.-Z.; Zou, H.-W.; Gicheha, M.G.; Wang, Z.-S. Effects of yeast and yeast cell wall polysaccharides supplementation on beef cattle growth performance, rumen microbial populations and lipopolysaccharides production. J. Integr. Agric. 2020, 19, 810–819. [Google Scholar] [CrossRef]
- Dias, A.L.G.; Freitas, J.A.; Micai, B.; Azevedo, R.A.; Greco, L.F.; Santos, J.E.P. Effects of supplementing yeast culture to diets differing in starch content on rumen fermentation and digestion in dairy cow. J. Dairy Sci. 2018, 101, 201–221. [Google Scholar] [CrossRef]
- Halfen, J.; Carpinelli, N.; Del Pino, F.; Chapman, J.; Sharman, E.; Anderson, J.; Osorio, J. Effects of yeast culture supplementation on lactation performance and rumen fermentation profile and microbial abundance in mid-lactation Holstein dairy cows. J. Dairy Sci. 2021, 104, 11580–11592. [Google Scholar] [CrossRef]
- Shen, J.; Zheng, L.; Chen, X.; Han, X.; Cao, Y.; Yao, J. Metagenomic Analyses of Microbial and Carbohydrate-Active Enzymes in the Rumen of Dairy Goats Fed Different Rumen Degradable Starch. Front. Microbiol. 2020, 11, 1003. [Google Scholar] [CrossRef]
- Gozho, G.N.; Plaizier, J.C.; Krause, D.O.; Kennedy, A.D.; Wittenberg, K.M. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an Inflammatory Response. J. Dairy Sci. 2005, 88, 1399–1403. [Google Scholar] [CrossRef] [Green Version]
- Pinart, M.; Dötsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut Microbiome Composition in Obese and Non-Obese Persons: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef]
- Myer, P.R.; Smith, T.P.L.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C. Rumen Microbiome from Steers Differing in Feed Efficiency. PLoS ONE 2015, 10, e0129174. [Google Scholar] [CrossRef]
- Meale, S.; Li, S.C.; Azevedo, P.; Derakhshani, H.; Devries, T.J.; Plaizier, J.C.; Steele, M.A.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Li, C.; Li, F.; Wang, X.; Zhang, X.; Liu, T.; Nian, F.; Yue, X.; Li, F.; Pan, X.; et al. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. Sci. Rep. 2016, 6, 32479. [Google Scholar] [CrossRef] [PubMed]
- Ogunade, I.M.; Lay, J.; Andries, K.; McManus, C.J.; Bebe, F. Effects of live yeast on differential genetic and functional attributes of rumen microbiota in beef cattle. J. Anim. Sci. Biotechnol. 2019, 10, 68. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Villot, C.; Renaud, D.; Skidmore, A.; Chevaux, E.; Steele, M.; Guan, L.L. Linking perturbations to temporal changes in diversity, stability, and compositions of neonatal calf gut microbiota: Prediction of diarrhea. ISME J. 2020, 14, 2223–2235. [Google Scholar] [CrossRef] [PubMed]
- Mondot, S.; Kang, S.; Furet, J.P.; de Cárcer, D.A.; McSweeney, C.; Morrison, M.; Marteau, P.; Doré, J.; Leclerc, M. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm. Bowel Dis. 2011, 17, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Fomenky, B.E.; Chiquette, J.; Bissonnette, N.; Talbot, G.; Chouinard, P.Y.; Ibeagha-Awemu, E.M. Impact of Saccharomyces cerevisiae boulardii CNCMI-1079 and Lactobacillus acidophilus BT1386 on total lactobacilli population in the gastrointestinal tract and colon histomorphology of Holstein dairy calves. Anim. Feed Sci. Technol. 2017, 234, 151–161. [Google Scholar] [CrossRef]
- Conlon, M.A.; Bird, A.R. The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef]
- Welty, C.; Wenner, B.; Wagner, B.; Roman-Garcia, Y.; Plank, J.; Meller, R.; Gehman, A.; Firkins, J. Rumen microbial responses to supplemental nitrate. II. Potential interactions with live yeast culture on the prokaryotic community and methanogenesis in continuous culture. J. Dairy Sci. 2019, 102, 2217–2231. [Google Scholar] [CrossRef] [Green Version]
- Bach, A.; López-García, A.; González-Recio, O.; Elcoso, G.; Fàbregas, F.; Chaucheyras-Durand, F.; Castex, M. Changes in the rumen and colon microbiota and effects of live yeast dietary supplementation during the transition from the dry period to lactation of dairy cows. J. Dairy Sci. 2019, 102, 6180–6198. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Zhang, N.; Wu, C.; Zhang, X.; Wang, Q.; Huang, X.; Du, L.; Cao, Q.; Tang, J.; Zhou, C.; et al. A metagenomic study of the gut microbiome in Behcet’s disease. Microbiome 2018, 6, 135. [Google Scholar] [CrossRef] [Green Version]
- Ogunade, I.; Schweickart, H.; Andries, K.; Lay, J.; Adeyemi, J. Monensin Alters the Functional and Metabolomic Profile of Rumen Microbiota in Beef Cattle. Animals 2018, 8, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, B.; Bach, A.; Guasch, I.; González, C.; Elcoso, G.; Pryce, J.E.; Gonzalez-Recio, O. Whole rumen metagenome sequencing allows classifying and predicting feed efficiency and intake levels in cattle. Sci. Rep. 2019, 9, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guedes, C.; Gonçalves, D.; Rodrigues, M.; Dias-Da-Silva, A. Effects of a Saccharomyces cerevisiae yeast on ruminal fermentation and fibre degradation of maize silages in cows. Anim. Feed Sci. Technol. 2008, 145, 27–40. [Google Scholar] [CrossRef]
- Patra, A.K.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Xue, M.-Y.; Xie, Y.-Y.; Zhong, Y.; Ma, X.-J.; Sun, H.-Z.; Liu, J.-X. Integrated meta-omics reveals new ruminal microbial features associated with feed efficiency in dairy cattle. Microbiome 2022, 10, 32. [Google Scholar] [CrossRef]
- Lynch, H.A.; Martin, S.A. Effects of Saccharomyces cerevisiae culture and Saccharomyces cerevisiae live cells on in vitro mixed ruminal microorganism fermentation. J. Dairy Sci. 2002, 85, 2603–2608. [Google Scholar] [CrossRef] [Green Version]
- Jin, D.; Kang, K.; Wang, H.; Wang, Z.; Xue, B.; Wang, L.; Xu, F.; Peng, Q. Effects of dietary supplementation of active dried yeast on fecal methanogenic archaea diversity in dairy cows. Anaerobe 2017, 44, 78–86. [Google Scholar] [CrossRef]
- Janssen, P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed. Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Denman, S.E.; Martinez Fernandez, G.; Shinkai, T.; Mitsumori, M.; McSweeney, C.S. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front. Microbiol. 2015, 6, 1087. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.F.; Bahram, M.; Han, J.-L.; Ding, X.-Z.; Salekdeh, G.H. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2020, 15, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, G.; Xu, H.; Xin, H.; Zhang, Y. Metagenomic Analyses of Microbial and Carbohydrate-Active Enzymes in the Rumen of Holstein Cows Fed Different Forage-to-Concentrate Ratios. Front. Microbiol. 2019, 10, 649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Hitch, T.C.A.; Chen, Y.; Creevey, C.; Guan, L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019, 7, 6. [Google Scholar] [CrossRef] [PubMed]


| Item | CON | LYP | HYP | SEM | p-Value |
|---|---|---|---|---|---|
| pH | 6.35 | 6.45 | 6.49 | 0.04 | 0.182 |
| Acetate, mmol/L | 50.11 | 49.16 | 57.16 | 3.04 | 0.086 |
| Propionate, mmol/L | 19.53 | 22.04 | 23.00 | 1.55 | 0.065 |
| Butytate, mmol/L | 15.66 b | 16.32 b | 19.65 a | 0.73 | <0.01 |
| TVFA, mmol/L | 85.30 b | 87.51 b | 99.81 a | 3.20 | 0.020 |
| Amylase, U/g.prot | 427.77 a | 379.87 b | 394.87 b | 5.60 | <0.01 |
| Cellulase, U/ml | 23.74 b | 26.07 b | 29.21 a | 0.83 | 0.011 |
| Ammonia, mg/dL | 12.55 | 11.19 | 11.10 | 0.89 | 0.565 |
| lactic acid, mmol/L | 5.46 a | 3.57 b | 3.12 b | 0.03 | <0.01 |
| LPS, KEU/ml | 43.85 a | 32.29 b | 21.73 c | 2.10 | <0.01 |
| HIS, ng/ml | 6.17 a | 3.71 b | 2.20 c | 0.05 | <0.01 |
| Item | CON | LYP | HYP | SEM | p-Value |
|---|---|---|---|---|---|
| Solid-associated Microorganism | |||||
| Methanobrevibacter | 44.78 a | 9.07 b | 11.95 b | 2.93 | <0.01 |
| Fusobacterium | 3.56 b | 5.40 b | 9.06 a | 0.46 | <0.01 |
| Dialister | 8.43 b | 111.52 a | 216.24 a | 20.17 | <0.01 |
| Ruminococcus | 389.02 a | 151.53 b | 406.60 a | 44.61 | 0.013 |
| Streptococcus | 4.78 b | 4.66 b | 5.61 a | 0.02 | 0.024 |
| Dorea | 3.59 b | 5.49 ab | 8.69 a | 0.87 | 0.038 |
| Lactobacillus | 6.33 b | 10.30 a | 9.18 a | 0.80 | 0.040 |
| Paraprevotella | 54.28 a | 16.65 b | 17.01 b | 1.95 | 0.047 |
| Liquid-associated Microorganism | |||||
| Mitsuokella | 17.65 b | 5.72 c | 42.14 a | 1.19 | <0.01 |
| Selenomonas | 110.45 b | 73.18 b | 276.76 a | 24.81 | 0.024 |
| Escherichia | 14.57 b | 40.83 a | 10.61 b | 3.38 | 0.026 |
| Megasphaera | 12.18 ab | 5.28 b | 19.56 a | 2.83 | 0.046 |
| Fusobacterium | 5.32 a | 2.85 b | 6.93 a | 0.82 | 0.048 |
| Genus ID | Description | CON | LYP | HYP | SEM | p-Values |
|---|---|---|---|---|---|---|
| Solid-associated microorganism | ||||||
| Streptococcus | Lactic acid-producing bacteria | 4.78 b | 4.66 b | 5.61 a | 0.16 | 0.024 |
| Butyrivibrio | 53.80 | 55.77 | 54.92 | 8.92 | 0.989 | |
| Lactobacillus | 6.33 b | 10.30 a | 9.18 a | 0.80 | 0.040 | |
| Mitsuokella | 13.47 | 31.96 | 19.14 | 7.60 | 0.310 | |
| Megasphaera | Lactic acid-utilizing bacteria | 8.58 | 20.29 | 22.37 | 3.25 | 0.051 |
| Selenomonas | 55.47 | 204.30 | 102.50 | 41.17 | 0.127 | |
| Fusobacterium | 3.56 b | 5.40 b | 9.06 a | 0.46 | <0.01 | |
| Veillonella | 1.85 | 2.70 | 5.28 | 0.71 | 0.084 | |
| Propionibacterium | 0.07 | 0.05 | 0.05 | 0.02 | 0.883 | |
| Liquid-associated microorganism | ||||||
| Streptococcus | Lactic acid-producing bacteria | 4.60 | 3.96 | 4.99 | 0.26 | 0.077 |
| Butyrivibrio | 35.37 | 27.42 | 34.04 | 5.96 | 0.666 | |
| Lactobacillus | 7.52 | 6.65 | 9.11 | 1.59 | 0.599 | |
| Mitsuokella | 17.65 b | 5.72 b | 42.14 a | 4.27 | <0.01 | |
| Megasphaera | Lactic acid-utilizing bacteria | 12.18 ab | 5.28 b | 19.56 a | 2.83 | 0.046 |
| Selenomonas | 110.45 b | 73.18 b | 276.76 a | 38.32 | 0.024 | |
| Fusobacterium | 5.32 b | 2.85 b | 6.93 a | 0.82 | 0.048 | |
| Veillonella | 2.10 b | 1.08 b | 3.37 a | 0.40 | 0.041 | |
| Propionibacterium | 0.14 | 0.12 | 0.03 | 0.06 | 0.598 | |
| Genus ID | CON | LYP | HYP | SEM | p-Value |
|---|---|---|---|---|---|
| Solid-associated microorganism | |||||
| Methanobrevibacter | 44.78 | 9.07 | 11.95 | 2.93 | <0.01 |
| Methanohalophilus | 0.02 | 0.02 | 0.08 | 0.01 | <0.01 |
| Methanosphaera | 1.15 | 0.37 | 0.52 | 0.08 | <0.01 |
| Methanomethylovorans | 0.02 | 0 | 0 | 0.00 | <0.01 |
| Methanoculleus | 0.1 | 0.04 | 0.02 | 0.01 | <0.01 |
| Methanoplanus | 0.08 | 0 | 0.01 | 0.01 | 0.015 |
| Methanothrix | 0.1 | 0.05 | 0.02 | 0.01 | 0.080 |
| Liquid-associated microorganism | |||||
| Methanothrix | 0.02 | 0.07 | 0.01 | 0.01 | 0.064 |
| Methanobrevibacter | 9.65 | 35.14 | 12.65 | 4.65 | 0.070 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.; Wang, H.; Shi, H.; Li, Q.; Zhang, Y.; Li, T.; Ma, Y. Yeast Products Mediated Ruminal Subenvironmental Microbiota, and Abnormal Metabolites and Digestive Enzymes Regulated Rumen Fermentation Function in Sheep. Animals 2022, 12, 3221. https://doi.org/10.3390/ani12223221
Su M, Wang H, Shi H, Li Q, Zhang Y, Li T, Ma Y. Yeast Products Mediated Ruminal Subenvironmental Microbiota, and Abnormal Metabolites and Digestive Enzymes Regulated Rumen Fermentation Function in Sheep. Animals. 2022; 12(22):3221. https://doi.org/10.3390/ani12223221
Chicago/Turabian StyleSu, Manchun, Huihui Wang, Huibin Shi, Qiao Li, Yong Zhang, Taotao Li, and Youji Ma. 2022. "Yeast Products Mediated Ruminal Subenvironmental Microbiota, and Abnormal Metabolites and Digestive Enzymes Regulated Rumen Fermentation Function in Sheep" Animals 12, no. 22: 3221. https://doi.org/10.3390/ani12223221
APA StyleSu, M., Wang, H., Shi, H., Li, Q., Zhang, Y., Li, T., & Ma, Y. (2022). Yeast Products Mediated Ruminal Subenvironmental Microbiota, and Abnormal Metabolites and Digestive Enzymes Regulated Rumen Fermentation Function in Sheep. Animals, 12(22), 3221. https://doi.org/10.3390/ani12223221








