Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Group Test and Sample Collection
- A.
- Free-range Gushi chicken without an additive group;
- B.
- Factory-bred Gushi chicken oregano oil group (0.25 mL/kg BW);
- C.
- Factory-bred Gushi chicken ENR group (5.0 mg/kg BW);
- D.
- Free-range Huainan pigs without an additive group;
- E.
- Factory-bred Huainan pigs without an additive group;
- F.
- Factory-bred Huainan pigs (2021011803). All samples were stored in the sample preservation solution, mixed with pig oregano oil group (0.25 mL/kg BW);
- G.
- Factory-bred Huainan pigs ENR Group (5.0 mg/kg BW)
- H.
- Factory-bred Gushi chicken without an additive group.
2.2. DNA Sample Detection
2.3. Library Construction and Library Inspection
2.4. On-Board Sequencing
2.5. Information Analysis Process
2.5.1. Data Quality Control
2.5.2. Metagenome Assembly
2.5.3. Gene Prediction
2.5.4. Species Annotation
2.5.5. Resistance Gene Annotation
3. Results
3.1. Sequencing Data Preprocessing Results
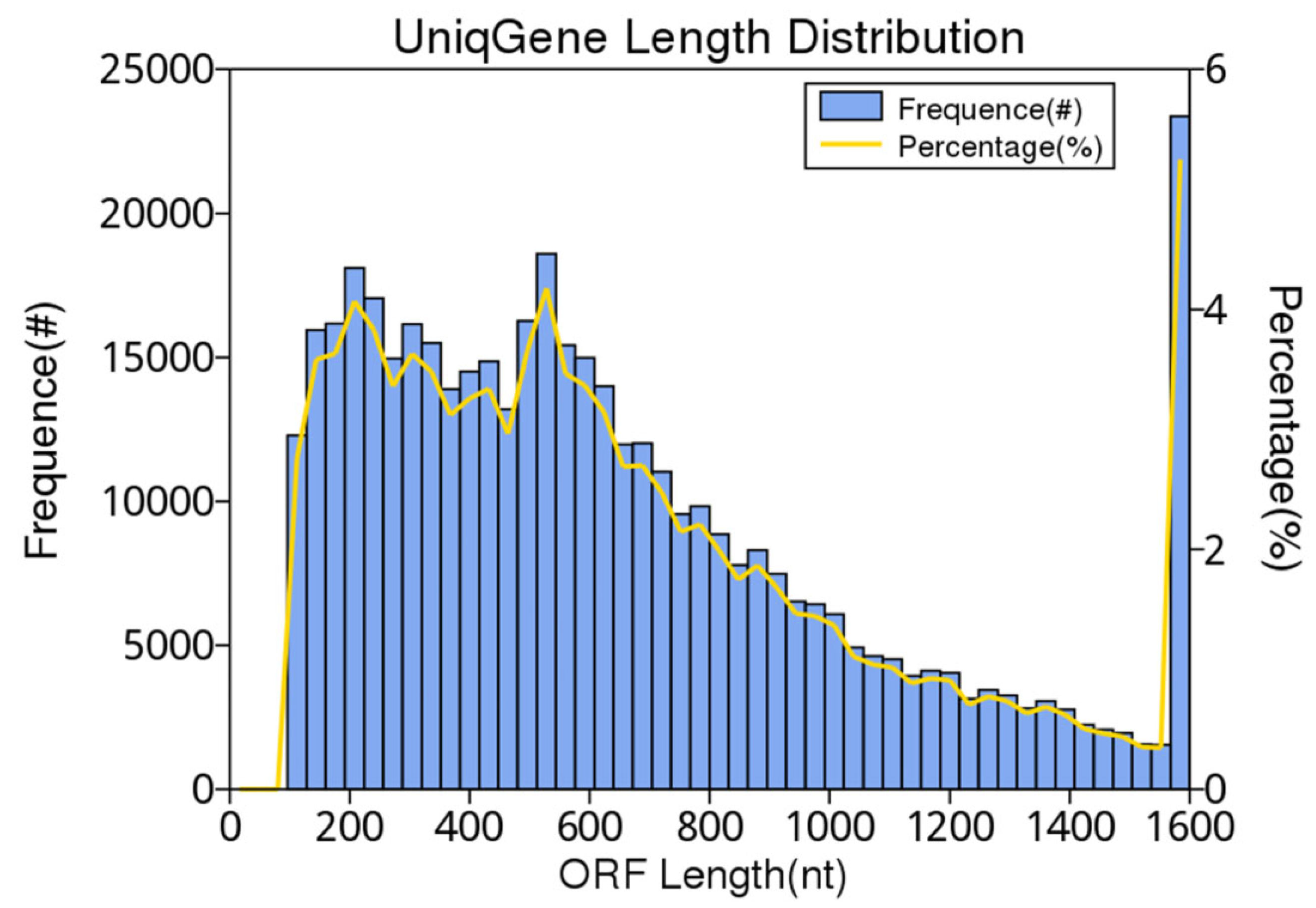
3.2. Metagenome Assembly (≥500 bp)
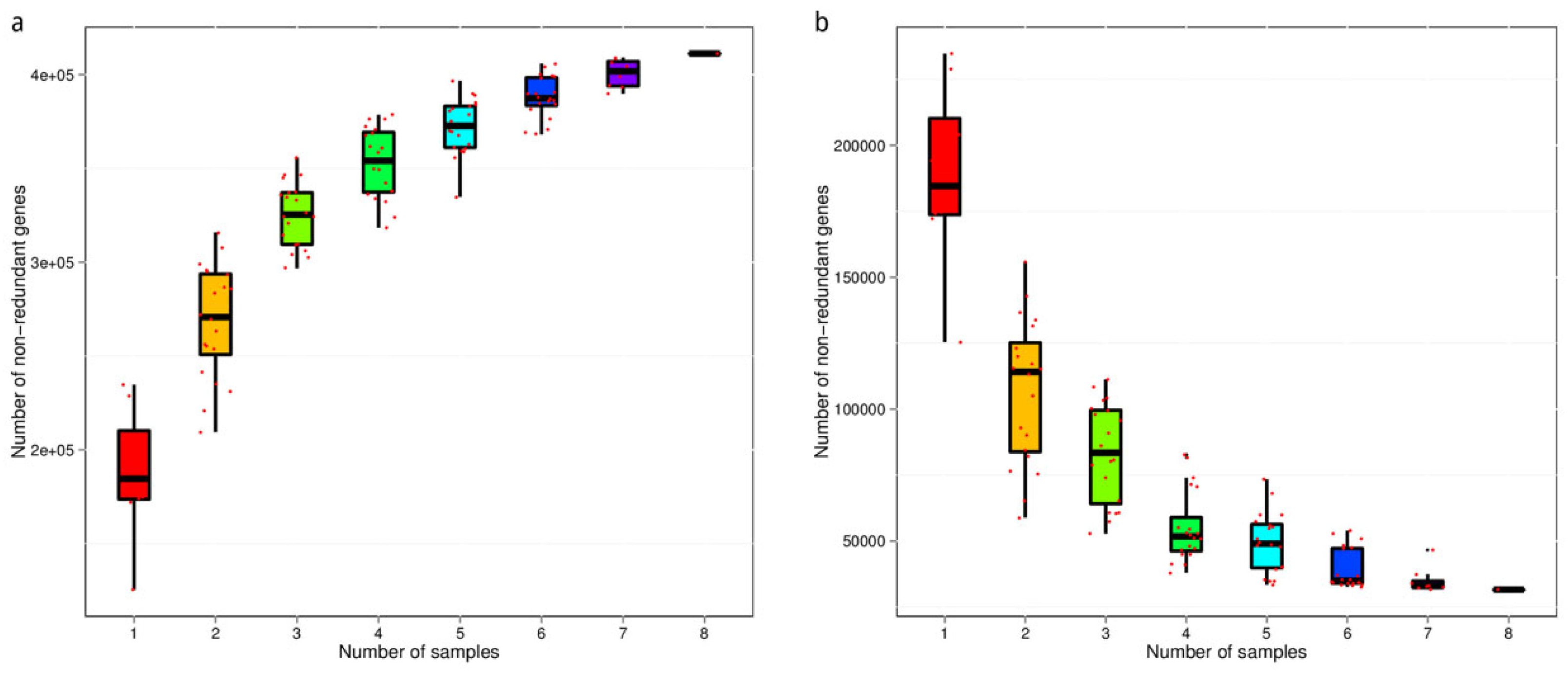
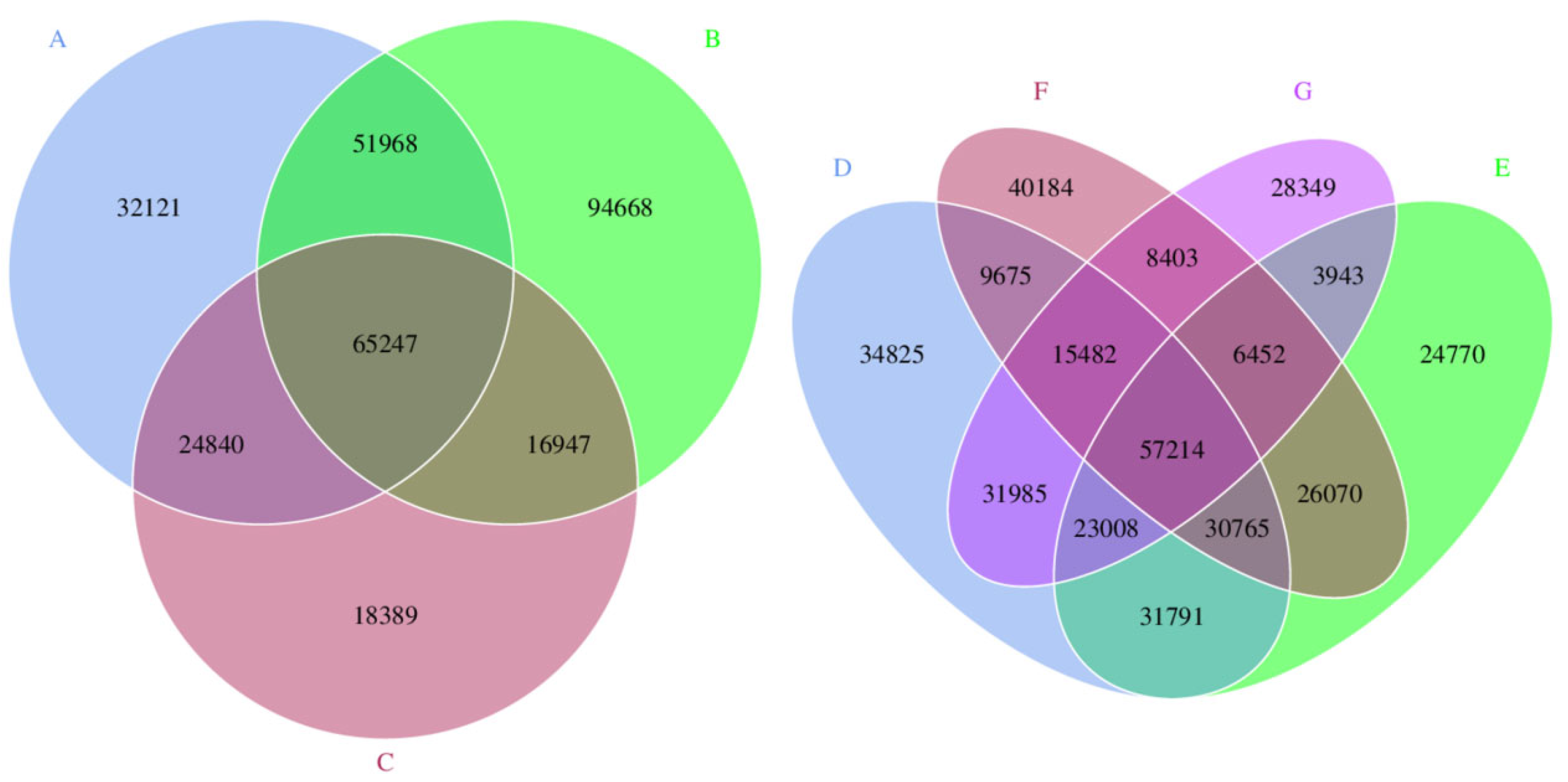
3.3. Core–Pan Gene Analysis
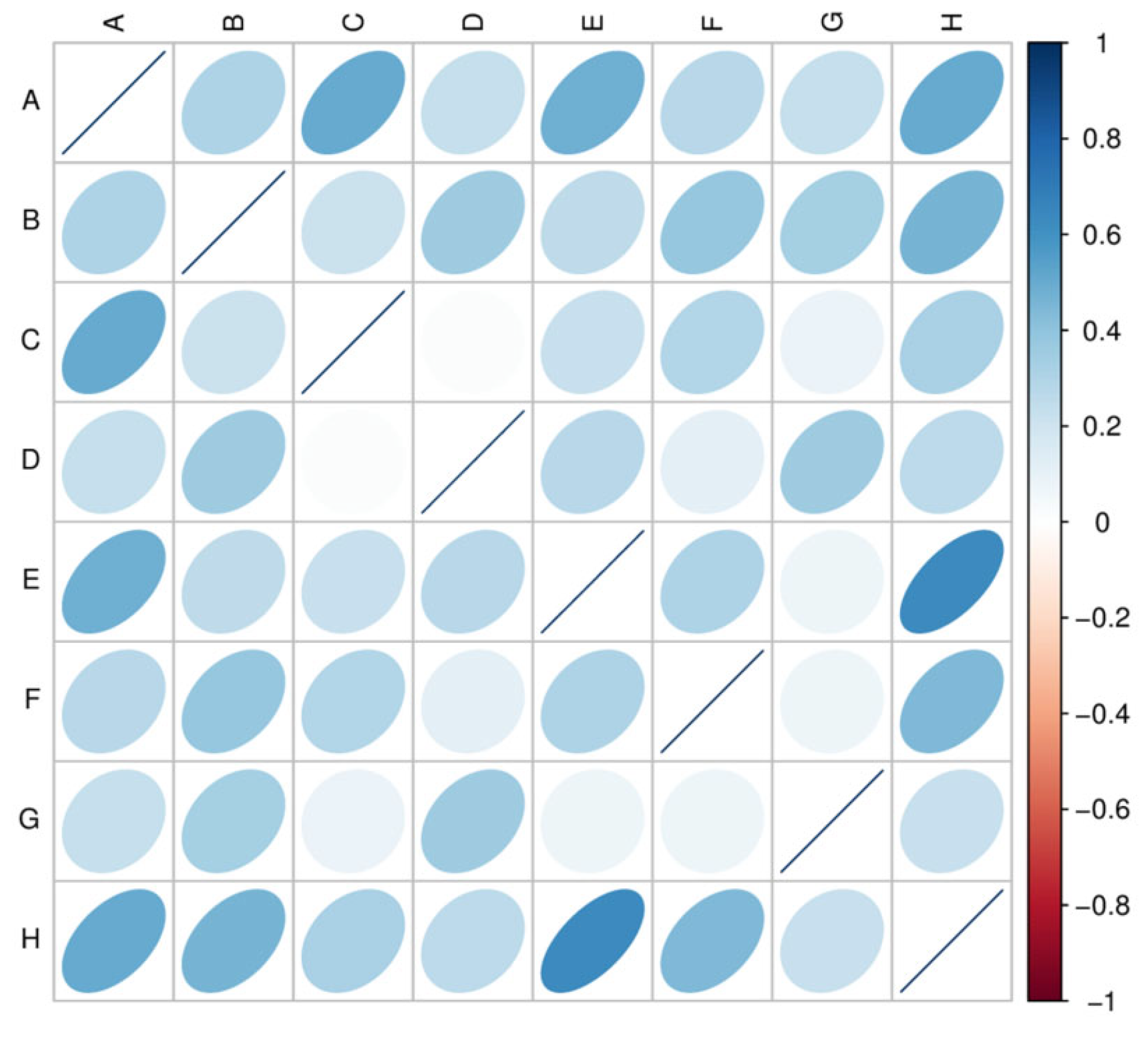
3.4. Gene Number-Based Correlation Analysis between Samples
3.5. Microbial Species Annotation
3.5.1. Basic Steps for Species Annotation
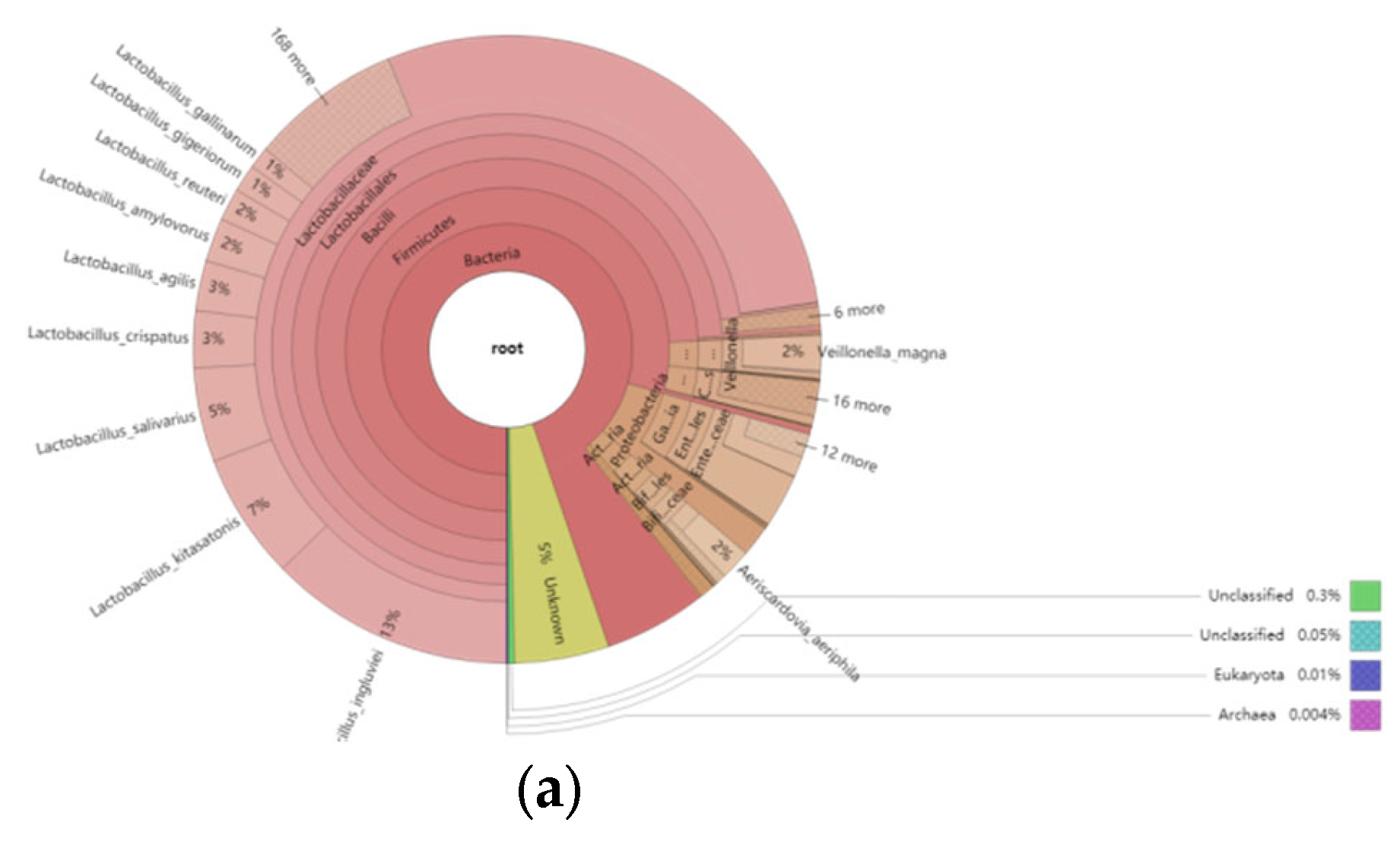
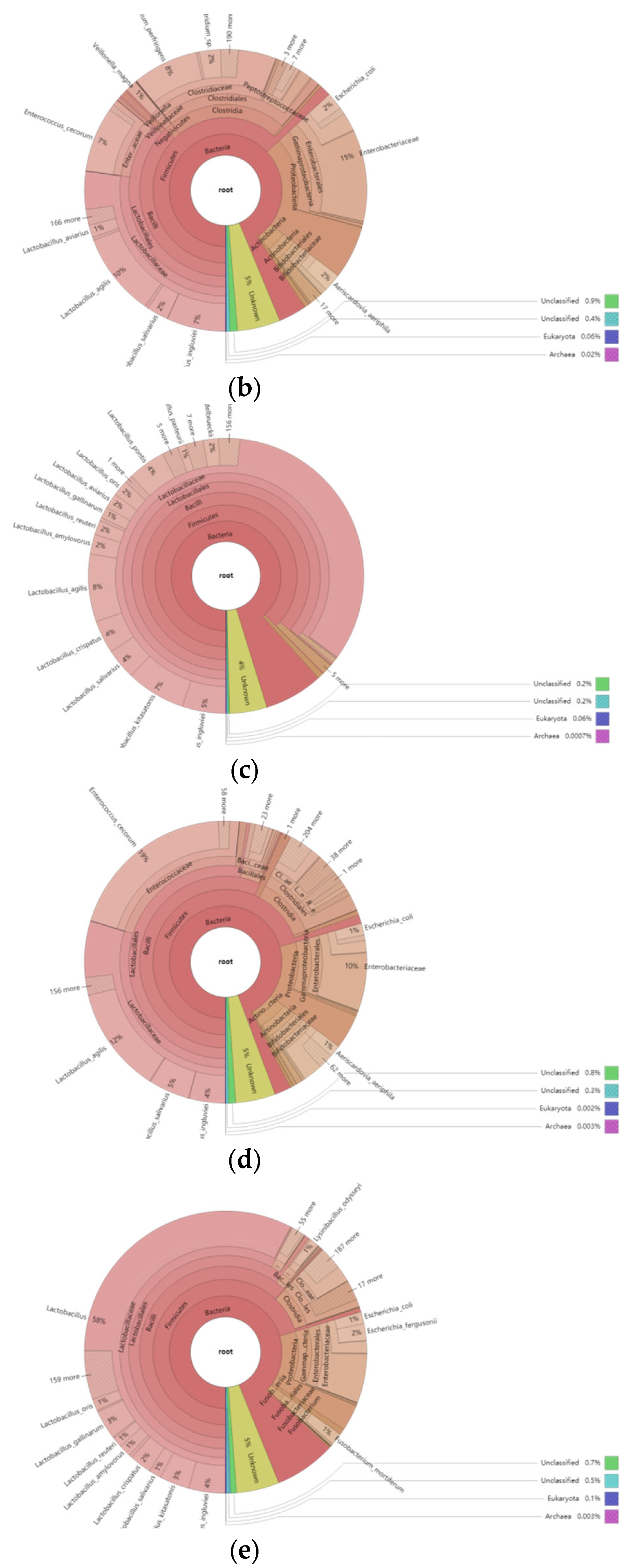
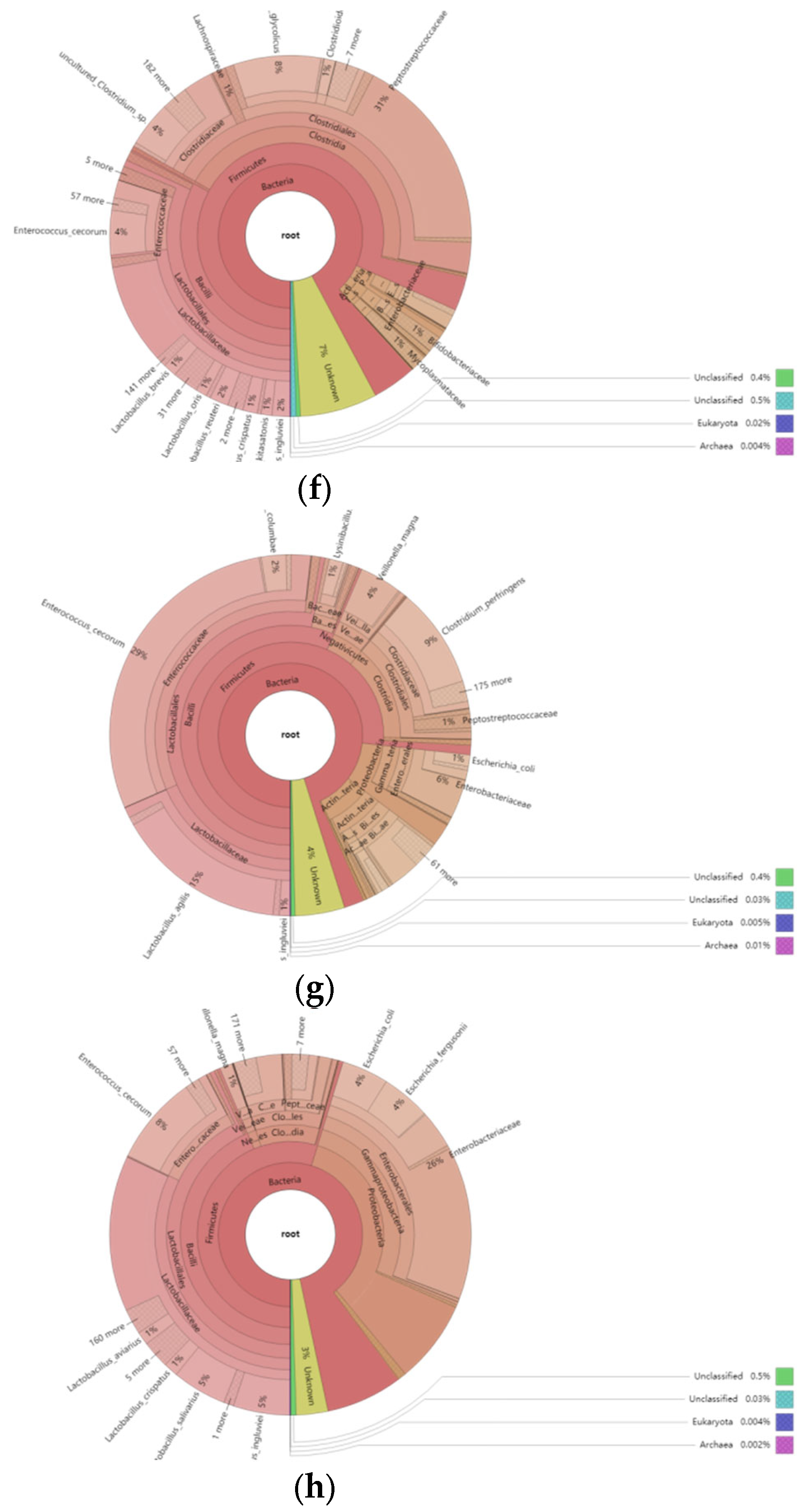
3.5.2. Overview of the Relative Abundance of Species
3.5.3. Cluster Analysis of the Number and Relative Abundance of Annotated Genes
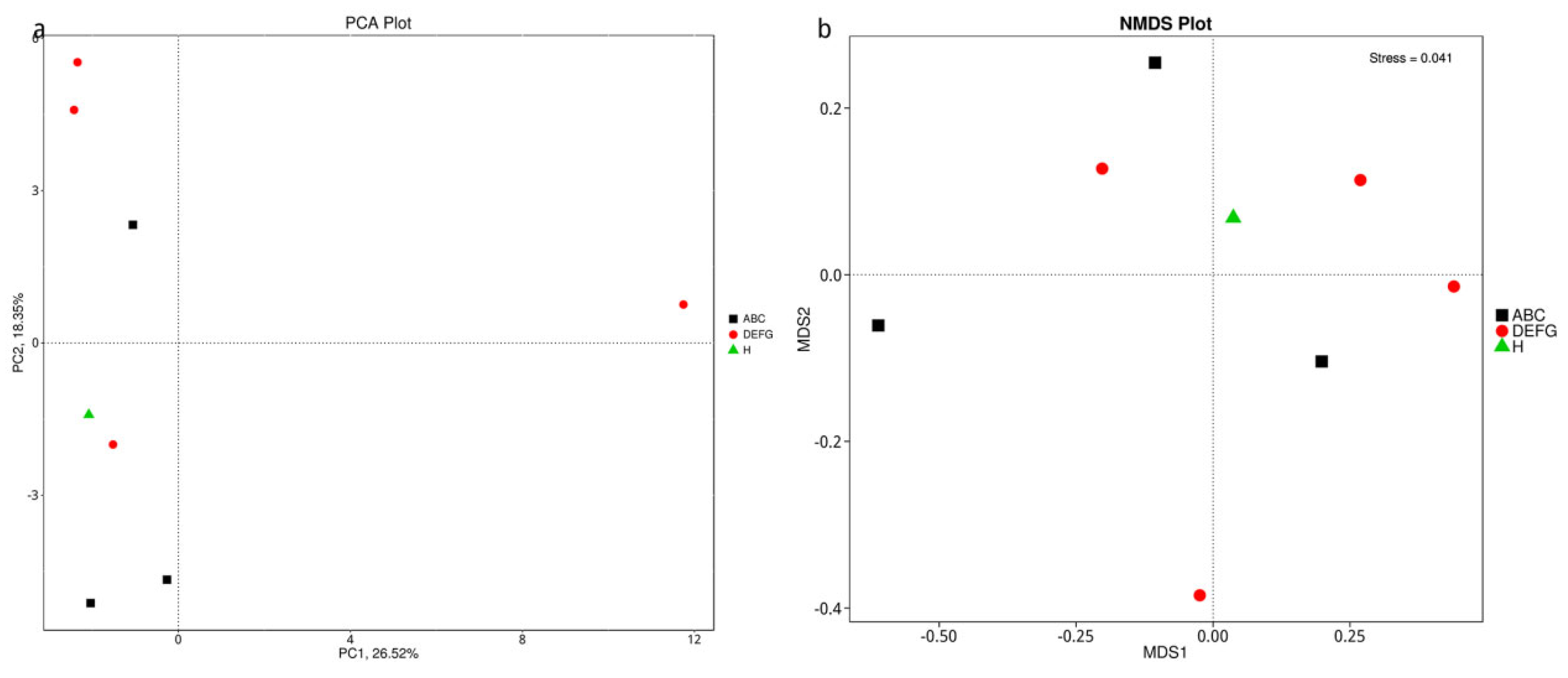
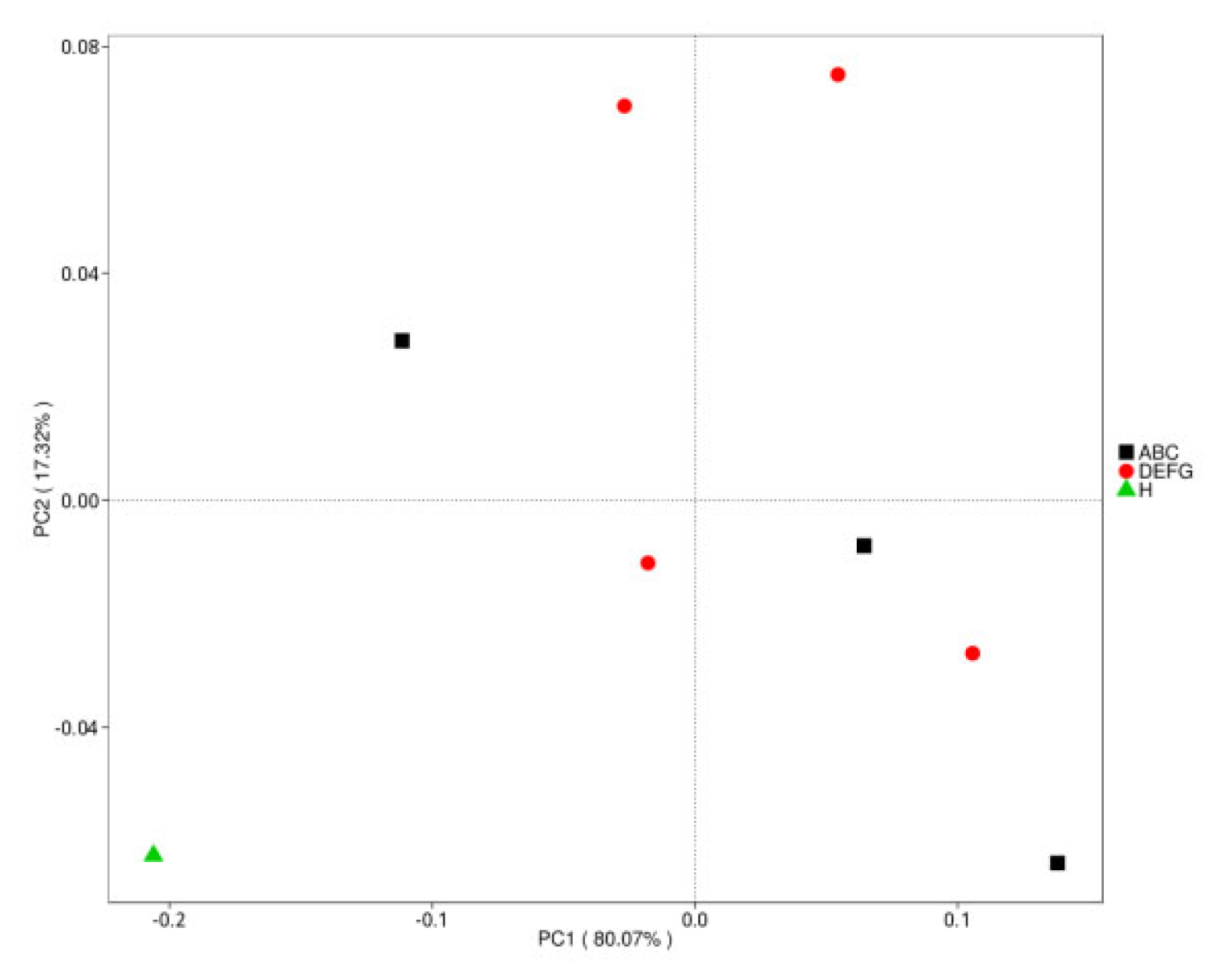
3.5.4. Dimension Reduction Analysis Based on Species Abundance
3.5.5. Dimension Reduction Analysis of Bray–Curtis Distance Based on Species Abundance
3.6. Cluster Analysis of Samples Based on Species Abundance
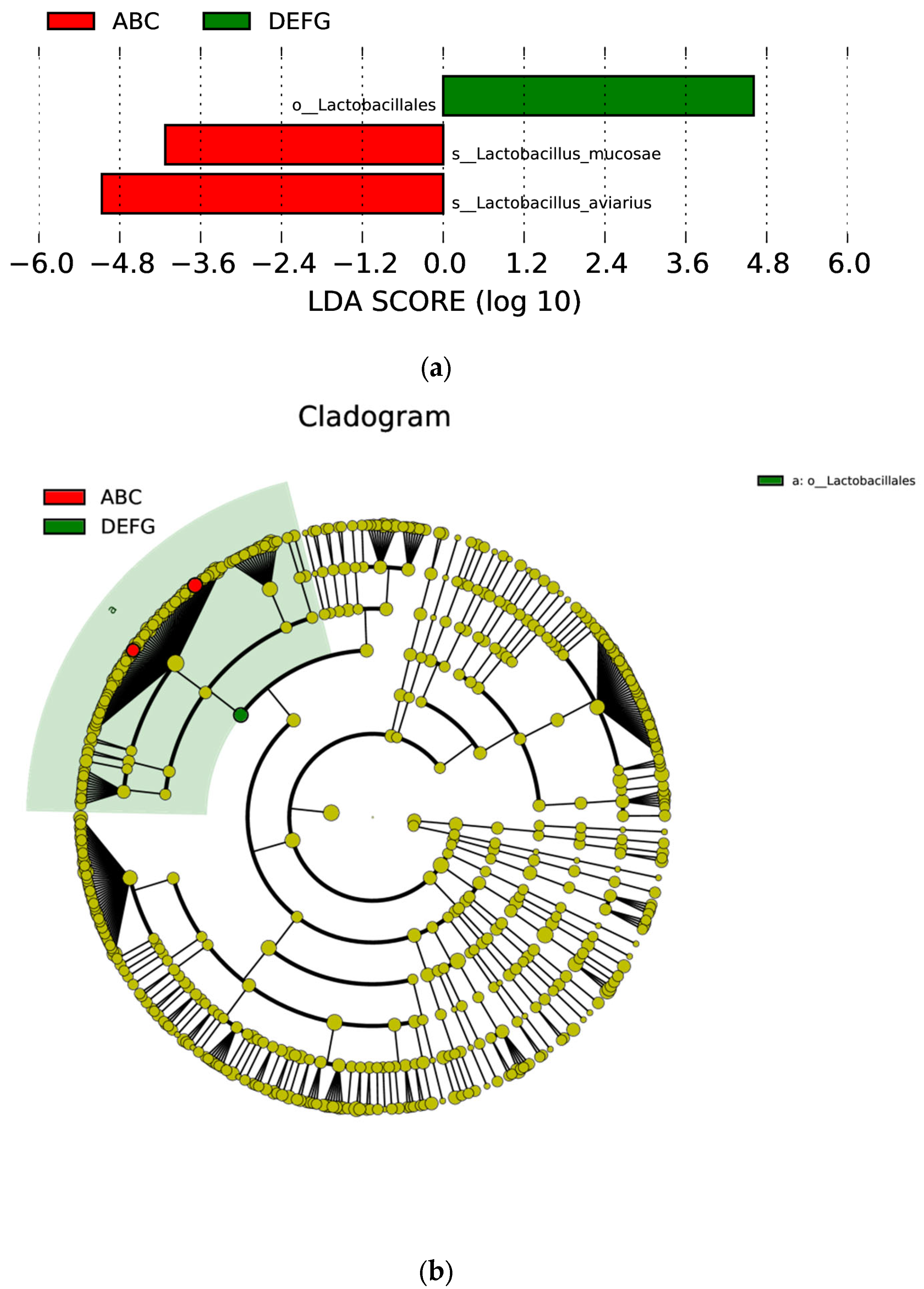
3.7. Linear Discriminant Analysis Effect Size (LEfSe) Analysis of Different Species between Groups
3.8. Annotation of Resistance Genes
3.8.1. Basic Steps of Resistance Gene Annotation
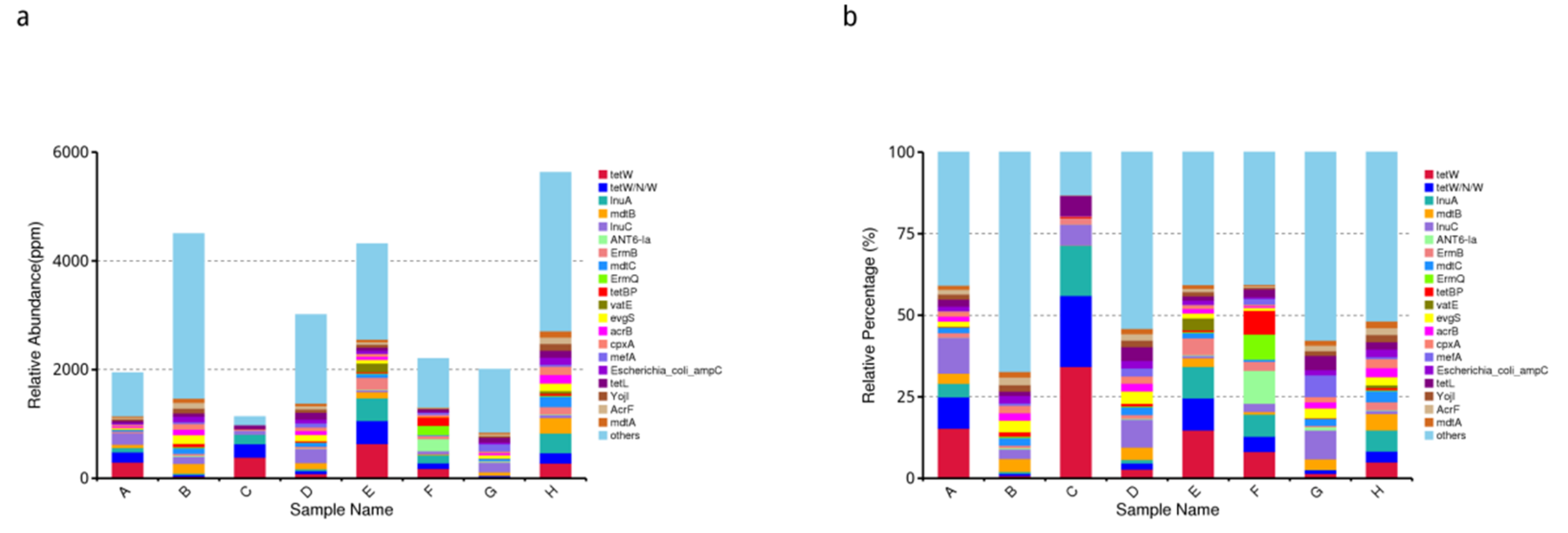
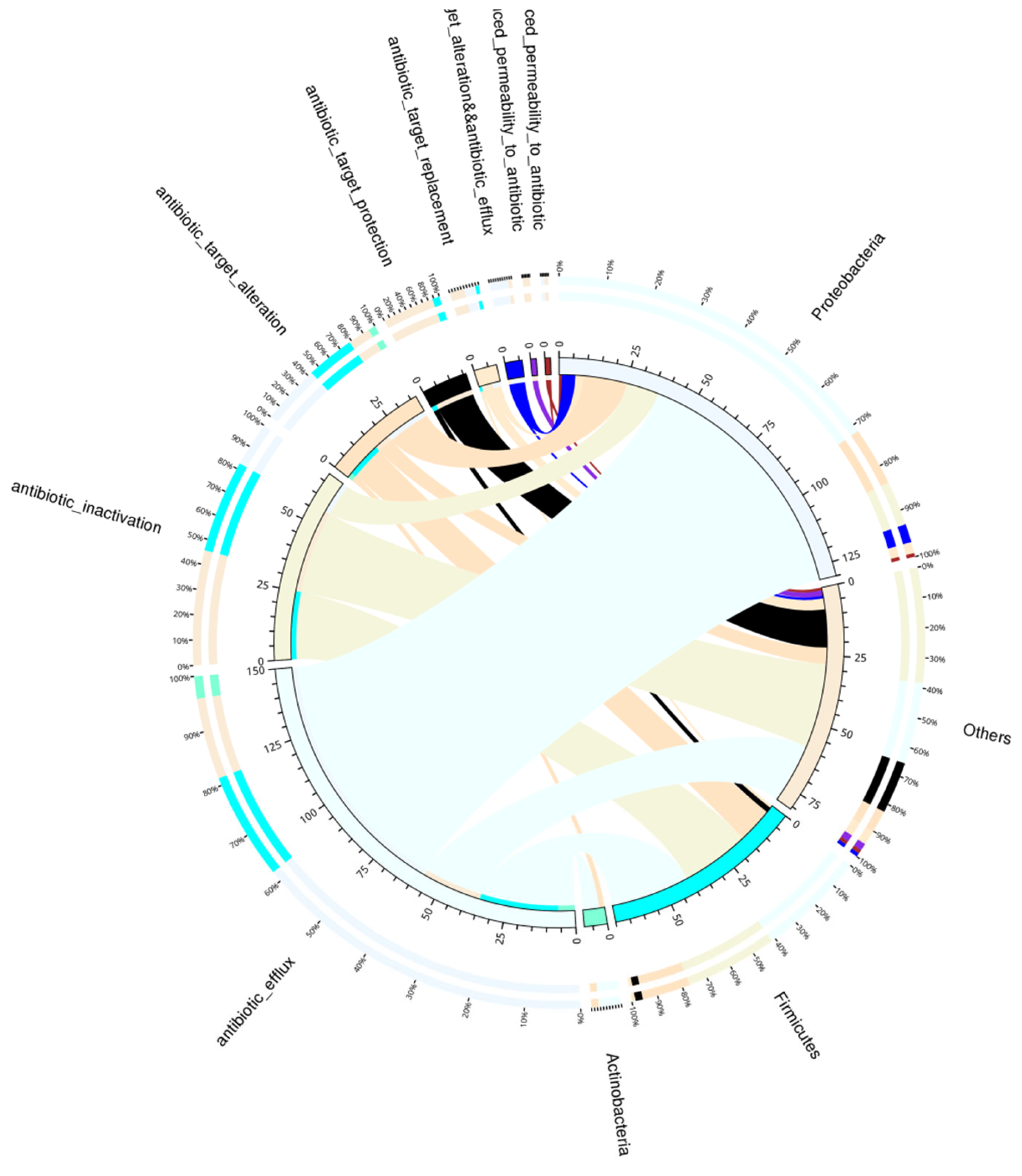
3.8.2. Overview of Resistance Gene Abundance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rustam, A. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2017, 133, 4–19. [Google Scholar]
- Sandelin, A.; Hälli, O.; Härtel, H.; Herva, T.; Kaartinen, L.; Tuunainen, E.; Rautala, H.; Soveri, T.; Simojoki, H. Effect of Farm Management Practices on Morbidity and Antibiotic Usage on Calf Rearing Farms. Antibiotics 2022, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Gnanadhas, D.P.; Marathe, S.A.; Chakravortty, D. Biocides-resistance, cross-resistance mechanisms and assessment. Expert Opin. Investig. Drugs 2013, 22, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, M.; Kundig, C. Microbial multidrug resistance. Int. J. Antimicrob. Agents 1997, 8, 179–187. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed]
- Besharati, S.; Motamedi, H.; Zallaghi, R. A survey on microbial quality and antibiotic resistance in Karoon River, Khuzestan, Iran. Appl. Water Sci. 2018, 8, 146. [Google Scholar] [CrossRef]
- Chiș, A.A.; Rus, L.L.; Morgovan, C.; Arseniu, A.M.; Frum, A.; Vonica-Țincu, A.L.; Gligor, F.G.; Mureșan, M.L.; Dobrea, C.M. Microbial Resistance to Antibiotics and Effective Antibiotherapy. Biomedicines 2022, 10, 1121. [Google Scholar] [CrossRef]
- Cloeckaert, A.; Schwarz, S. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica Typhimurium DT104. Vet. Res. 2001, 32, 301–310. [Google Scholar] [CrossRef]
- Erol, E.; Scortti, M.; Fortner, J.; Patel, M.; Vázquez-Boland, J.A. Antimicrobial Resistance Spectrum Conferred by pRErm46 of Emerging Macrolide (Multidrug)-Resistant Rhodococcus equi. J. Clin. Microbiol. 2021, 59, e0114921. [Google Scholar] [CrossRef]
- Getahun, Y.A.; Ali, D.A.; Taye, B.W.; Alemayehu, Y.A. Multidrug-Resistant Microbial Therapy Using Antimicrobial Peptides and the CRISPR/Cas9 System. Veter.-Med. Res. Rep. 2022, 13, 173–190. [Google Scholar] [CrossRef]
- Khare, T.; Anand, U.; Dey, A.; Assaraf, Y.G.; Chen, Z.S.; Liu, Z.; Kumar, V. Exploring Phytochemicals for Combating Antibiotic Resistance in Microbial Pathogens. Front. Pharmacol. 2021, 12, 720–726. [Google Scholar]
- Liu, W.; Liu, Y.T. Roles of Multidrug Resistance Protein 4 in Microbial Infections and Inflammatory Diseases. Microb. Drug Resist. 2021, 27, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.C.; Liao, W.Y.; Chou, M.C.; Wu, Y.Y.; Yeh, T.H.; Lo, H.R. Overexpression of Resistance-Nodulation-Division Efflux Pump Genes Contributes to Multidrug Resistance in Aeromonas hydrophila Clinical Isolates. Microb. Drug Resist. 2022, 28, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Souza, K.L.D.; Fuzatti, J.V.S.; Camargo, R.C.; Pinto, M.D.; Silva, T.K.; Frias, D.F.R. Prevalence of multidrug-resistant bacteria in the nasal cavity of horses asymptomatic for respiratory diseases. Rev. Univap. 2020, 26, 107–123. [Google Scholar] [CrossRef]
- Van Veen, H.W.; Konings, W.N. The ABC family of multidrug transporters in microorganisms. Biochim. Biophys. Acta (BBA)—Bioenerg. 1998, 1365, 31–36. [Google Scholar] [CrossRef]
- Yewale, P.P.; Lokhande, K.B.; Sridhar, A.; Vaishnav, M.; Khan, F.A.; Mandal, A.; Swamy, K.V.; Jass, J.; Nawani, N. Molecular profiling of multidrug-resistant river water isolates: Insights into resistance mechanism and potential inhibitors. Environ. Sci. Pollut. Res. 2020, 27, 27279–27292. [Google Scholar] [CrossRef]
- Sun, K.; Zhang, J.M.; Jiang, D.W.; Wang, X.; Ge, Y.; Deng, X. Progress and Countermeasures of Antimicrobial Resistance of Animal Origin Bacterial Pathogens in China. J. Agric. Sci. Technol. 2020, 22, 1–5. [Google Scholar]
- Chen, C.; Zhou, Y.; Fu, H.; Xiong, X.; Fang, S.; Jiang, H.; Wu, J.; Yang, H.; Gao, J.; Huang, L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021, 12, 1106. [Google Scholar] [CrossRef]
- Zhou, Y.Y. Construction and Application of the Catalogs of Microbial Genomes, Genes and Antimicrobial Resistance Genes from Pig Gut Microbiome; Jiangxi Agricultural University: Nanchang, China, 2021. [Google Scholar]
- Lu, Z.; Shen, Q.C.; Zhang, C.P.; Qi, Z. Predictive analysis of whole genome sequencing for Salmonella serotype and antimicrobial resistance phenotypes. Acta Microbiol. Sin. 2021, 61, 4038–4047. [Google Scholar]
- Chen, K.; Pachter, L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005, 1, 106–112. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [PubMed]
- Tringe, S.G.; Rubin, E.M. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005, 6, 805–814. [Google Scholar] [CrossRef]
- Tringe, S.G.; von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Foerstner, K.U.; Bork, P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr. Opin. Microbiol. 2007, 10, 490–498. [Google Scholar] [CrossRef]
- Liu, P.; Jiang, Y.; Gu, S.; Xue, Y.; Yang, H.; Li, Y.; Wang, Y.; Yan, C.; Jia, P.; Lin, X.; et al. Metagenome-wide association study of gut microbiome revealed potential microbial marker set for diagnosis of pediatric myasthenia gravis. BMC Med. 2021, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hao, H.; Cheng, G.; Liu, C.; Ahmed, S.; Shabbir, M.A.; Hussain, H.I.; Dai, M.; Yuan, Z. Microbial shifts in the intestinal microbiota of Salmonella infected chickens in response to enrofloxacin. Front. Microbiol. 2017, 8, 1711. [Google Scholar] [CrossRef]
- De Smet, J.; Boyen, F.; Croubels, S.; Rasschaert, G.; Haesebrouck, F.; Temmerman, R.; Rutjens, S.; De Backer, P.; Devreese, M. The impact of therapeutic-dose induced intestinal enrofloxacin concentrations in healthy pigs on fecal Escherichia coli populations. BMC Vet. Res. 2020, 16, 382. [Google Scholar] [CrossRef]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef]
- Wang, C.; Li, P.; Yan, Q.; Chen, L.; Li, T.; Zhang, W.; Li, H.; Chen, C.; Han, X.; Zhang, S.; et al. Characterization of the Pig Gut Microbiome and Antibiotic Resistome in Industrialized Feedlots in China. mSystems 2019, 4, e00206-19. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Bao, Y.; Zhang, Z.; Zhao, W.; Xiao, J.; He, S.; Zhang, G.; Li, Y.; Zhao, G.; Chen, R.; et al. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [Google Scholar]
- Wen, Y.; Li, S.; Wang, Z.; Feng, H.; Yao, X.; Liu, M.; Chang, J.; Ding, X.; Zhao, H.; Ma, W. Intestinal Microbial Diversity of Free-Range and Captive Yak in Qinghai Province. Microorganisms 2022, 10, 754. [Google Scholar] [CrossRef]





| #Sample | InsertSize (bp) | RawData | CleanData | Clean_Q20 | Clean_Q30 | Clean_GC (%) | Effective (%) |
|---|---|---|---|---|---|---|---|
| A | 350 | 6633.18 | 6629.30 | 96.2 | 89.95 | 40.72 | 99.941 |
| B | 350 | 6982.84 | 6969.06 | 96.11 | 89.8 | 41.56 | 99.803 |
| C | 350 | 6714.52 | 6708.95 | 96.27 | 90.09 | 40.08 | 99.917 |
| D | 350 | 6155.96 | 6152.45 | 96.03 | 89.66 | 41.1 | 99.943 |
| E | 350 | 6822.24 | 6818.75 | 95.88 | 89.34 | 39.31 | 99.949 |
| F | 350 | 6528.38 | 6524.90 | 95.99 | 89.38 | 35.21 | 99.947 |
| G | 350 | 6884.02 | 6881.10 | 95.95 | 89.5 | 39.52 | 99.958 |
| H | 350 | 6645.17 | 6639.25 | 96.39 | 90.44 | 42.14 | 99.911 |
| Sample ID | Total Len. (bp) | No. | Average Len. (bp) | N50 Len. (bp) | N90 Len. (bp) | Max Len. (bp) |
|---|---|---|---|---|---|---|
| A | 77,097,982 | 50,858 | 1515.95 | 2087 | 625 | 304,611 |
| B | 103,806,670 | 79,760 | 1301.49 | 1565 | 598 | 94,301 |
| C | 53,150,756 | 34,873 | 1524.12 | 2127 | 628 | 182,450 |
| D | 114,767,070 | 76,524 | 1499.75 | 2000 | 634 | 269,282 |
| E | 91,187,139 | 64,129 | 1421.93 | 1804 | 619 | 395,129 |
| F | 98,440,700 | 58,234 | 1690.43 | 2689 | 644 | 185,631 |
| G | 85,667,957 | 51,678 | 1657.73 | 2510 | 637 | 297,861 |
| H | 77,519,231 | 54,238 | 1429.24 | 1830 | 617 | 157,857 |
| ORFs No. | 445,260 |
|---|---|
| integrity:end | 82,047 (18.43%) |
| integrity:start | 93,479 (20.99%) |
| integrity:all | 237,785 (53.4%) |
| integrity:none | 31,949 (7.18%) |
| Total Len. (Mbp) | 300.11 |
| Average Len. (bp) | 674.02 |
| GC percent | 39.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rui, Y.; Qiu, G. Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China. Animals 2022, 12, 3404. https://doi.org/10.3390/ani12233404
Rui Y, Qiu G. Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China. Animals. 2022; 12(23):3404. https://doi.org/10.3390/ani12233404
Chicago/Turabian StyleRui, Yapei, and Gang Qiu. 2022. "Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China" Animals 12, no. 23: 3404. https://doi.org/10.3390/ani12233404
APA StyleRui, Y., & Qiu, G. (2022). Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China. Animals, 12(23), 3404. https://doi.org/10.3390/ani12233404






