Genome-Wide Association Analysis Reveals Novel Loci Related with Visual Score Traits in Nellore Cattle Raised in Pasture–Based Systems
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypes, Genotypes, and Pedigree
2.2. Data Quality Control
2.3. Single-Step Genome-Wide Association Study (ssGWAS)
2.4. Estimation of SNP Effects
2.5. Identification of Candidate Genes and Functional Analyses
3. Results
3.1. Descriptive Statistics and Variance Components
3.2. Single-Step GWAS
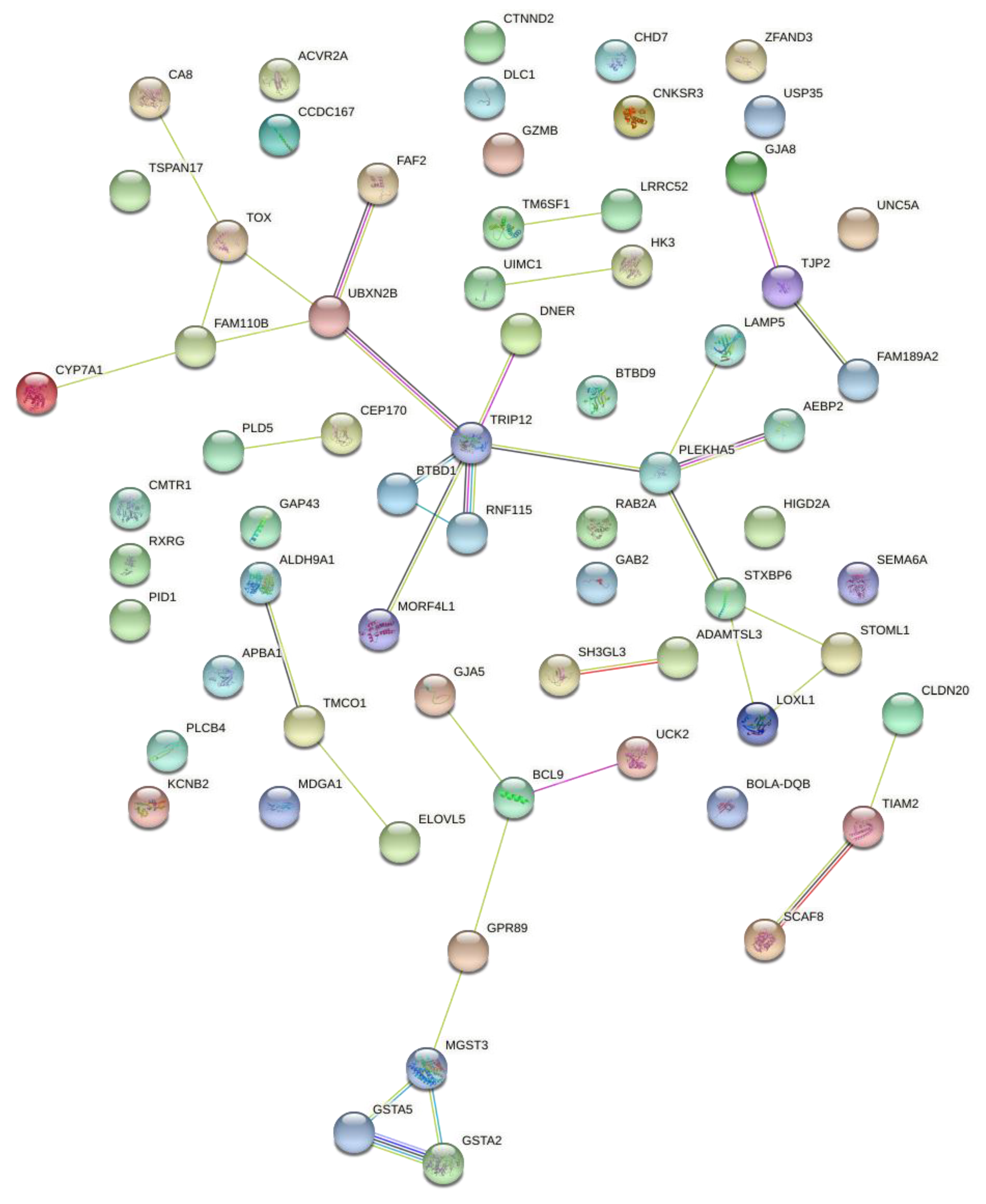
3.3. Functional Analyses and Gene Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abreu, L.R.A.; Martins, P.G.M.A.; Mota, L.F.M.; Ferreira, T.A.; Ribeiro, V.M.P.; Villela, S.D.J.; Merlo, F.A.; Pires, A.V. Genetic correlations between body weight, scrotal circumference and visual evaluation scores in Bos indicus cattle. Anim. Sci. J. 2015, 89, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, B.B.M.; MacNeil, M.D.; Da Costa, R.F.; Dionello, N.J.L.; Yokoo, M.J.; Cardoso, F.F. Genetic Parameters and Trends for Traits of the Hereford and Braford Breeds in Brazil. Livest. Sci. 2018, 208, 60–66. [Google Scholar] [CrossRef]
- Silva, R.P.; Espigolan, R.; Berton, M.P.; Lôbo, R.B.; Magnabosco, C.U.; Pereira, A.S.C.; Baldi, F. Genomic Prediction Ability for Carcass Composition Indicator Traits in Nellore Cattle. Livest. Sci. 2021, 245, 104421. [Google Scholar] [CrossRef]
- Vargas, G.; Schenkel, F.S.; Brito, L.F.; Neves, H.H.R.; Munari, D.P.; Boligon, A.A.; Carvalheiro, R. Unravelling biological biotypes for growth, visual score and reproductive traits in Nellore cattle via principal component analysis. Livest. Sci. 2018, 217, 37–43. [Google Scholar] [CrossRef]
- Souza, J.S.; Silveira, D.D.; Teixeira, B.B.M.; Boligon, A.A. Parameters and genetic associations of visual scores and weights in Hereford and Braford breeds. Livest. Sci. 2020, 241, 104216. [Google Scholar] [CrossRef]
- Boligon, A.A.; Farias, P.P.; Roso, V.M.; Santana, M.L.; Bignardi, A.B.; Souza, F.R.P. Genetic relations and indirect response to selection based on indices for scrotal circumference, visual scores and weight gain in beef cattle. Anim. Prod. Sci. 2018, 58, 1210–1217. [Google Scholar] [CrossRef]
- Silveira, D.D.; De Vargas, L.; Pereira, R.J.; Lôbo, R.B.; De Souza, F.R.P.; Boligon, A.A. Beef Cattle Growth Deceleration Parameters and Its Correlations with Growth, Carcass and Morphological Composition Traits. Livest. Sci. 2018, 214, 167–174. [Google Scholar] [CrossRef]
- Cancian, P.H.; Gomes, R.C.; Manicardi, F.R.; Ianni, A.C.; Bonin, M.N.; Leme, P.R.; Silva, S.R. Correlations of Visual Scores, Carcass Traits, Feed Efficiency and Retail Product Yield in Nellore Cattle. Sci. Agric. 2014, 71, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Stewart, S.M.; Gardner, G.E.; Williams, A.; Pethick, D.W.; McGilchrist, P.; Kuchida, K. Association between Visual Marbling Score and Chemical Intramuscular Fat with Camera Marbling Percentage in Australian Beef Carcasses. Meat Sci. 2020, 181, 108369. [Google Scholar] [CrossRef]
- Viana, A.F.P.; Rorato, P.R.N.; Mello, F.C.B.; Machado, D.S.; Figueiredo, A.M.; Bravo, A.P.; Feltes, G.L. Principal Component Analysis of Breeding Values for Growth, Reproductive and Visual Score Traits of Nellore Cattle. Livest. Sci. 2020, 241, 104262. [Google Scholar] [CrossRef]
- Martins, R.; Machado, P.C.; Pinto, L.F.B.; Silva, M.R.; Schenkel, F.S.; Brito, L.F.; Pedrosa, V.B. Genome-Wide Association Study and Pathway Analysis for Fat Deposition traits in Nellore Cattle Raised in Pasture–based Systems. J. Anim. Breed. Genet. 2020, 138, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Abdalla, E.A.; Maltecca, C.; Baes, C.F. Invited Review: Reproductive and Genomic Technologies to Optimize Breeding Strategies for Genetic Progress in Dairy Cattle. Arch. Anim. Breed. 2018, 61, 43–57. [Google Scholar] [CrossRef] [Green Version]
- Vanvanhossou, S.F.U.; Scheper, C.; Dossa, L.H.; Yin, T.; Brügemann, K.; König, S. A multi-breed GWAS for morphometric traits in four Beninese indigenous cattle breeds reveals loci associated with conformation, carcass and adaptive traits. BMC Genom. 2020, 21, 783. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Machado, P.C.; Pinto, L.F.B.; Silva, M.R.; Schenkel, F.S.; Brito, L.F.; Pedrosa, V.B. Genome-Wide Association Study and Pathway Analysis for Carcass Fatness in Nellore Cattle Raised in Pasture–based Systems. J. Anim. Genet. 2021, 52, 730–733. [Google Scholar] [CrossRef]
- Pedrosa, V.B.; Scenkel, F.S.; Che, S.Y.; Oliveira, H.R.; Casey, T.M.; Melka, M.G.; Brito, L.F. Genome-wide Association Analyses of Lactation Persistency and Milk Production Traits in Holstein Cattle Based on Imputed Whole-Genome Sequence Data Genes. Genes 2021, 12, 1830. [Google Scholar] [CrossRef]
- Naserkheil, M.; BahramI, A.; Lee, D.; Mehrban, H. Integrating Single–Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle. Animals 2020, 10, 1836. [Google Scholar] [CrossRef]
- Srikanth, K.; Lee, S.H.; Chung, K.Y.; Park, J.E.; Jang, G.W.; Park, M.R.; Kim, N.Y.; Kim, T.H.; Chai, H.H.; Park, W.C.; et al. A Gene-Set Enrichment and Protein–protein Interaction Network-Based Gwas with Regulatory Snps Identifies Candidate Genes and Pathways Associated with Carcass Traits in Hanwoo Cattle. Genes 2020, 11, 316. [Google Scholar] [CrossRef] [Green Version]
- Carreño, L.O.D.; Pessoa, M.C.; Espigolan, R.; Takada, L.; Bresolin, T.; Cavani, L.; Baldi, F.; Carvalheiro, R.; Albuquerque, L.G.; Fonseca, R. Genome Association Study for Visual Scores in Nellore Cattle Measured at Weaning. BMC Genom. 2019, 20, 150. [Google Scholar] [CrossRef] [Green Version]
- Silva, R.P.; Berton, M.P.; Grigoletto, L.; Carvalho, F.E.; Silva, R.M.O.; Peripolli, E.; Castro, L.M.; Ferraz, J.B.S.; Eler, J.P.; Lôbo, R.B.; et al. Genomic Regions and Enrichment Analyses Associated with Carcass Composition Indicator Traits in Nellore Cattle. J. Anim. Breed. Genet. 2019, 136, 118–133. [Google Scholar] [CrossRef]
- Sánchez, N.M.; Rueda, J.; Carabaño, M.J.; Reverter, A.; Mcwilliam, S.; González, C.; Díaz, C. Skeletal Muscle Specific Genes Networks in Cattle. Funct. Integr. Genom. 2010, 10, 609–618. [Google Scholar] [CrossRef]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef]
- Aiello, D.; Patel, K.; Lasagna, E. The myostatin gene: An overview of mechanisms of action and its relevance to livestock animals. Anim. Genet. 2018, 49, 505–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machugh, D.E.; Reecy, J.; Nolte, W.; Weikard, R.; Brunner, R.M.; Albrecht, E.; Hammon, H.M.; Reverter, A.; Kühn, C. Biological Network Approach for the Identification of Regulatory Long Non-Coding RNAs Associated with Metabolic Efficiency in Cattle. Front. Genet. 2019, 10, 1130. [Google Scholar] [CrossRef]
- Koury Filho, W.; Albuquerque, L.G.; Forni, S.; Silva, J.A.V.; Yokoo, M.J.; Alencar, M.M. Estimativas de parâmetros genéticos para os escores visuais e suas associações com peso corporal em bovinos de corte. Rev. Bras. Zootec. 2010, 39, 1015–1022. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.F.E.; Maniatis, T. Molecular cloning: A laboratory manual. Anal. Biochem. 1989, 170, 182–183. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, GA, USA, 2018; p. 125. [Google Scholar]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. Blupf90 and Related Programs (Bgf90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002. [Google Scholar]
- Aguilar, I.; Misztal, I.; Tsuruta, S.; Legarra, A. PREGSF90—POSTGSF90: Computational Tools for the Implementation of Single-step Genomic Selection and Genome-Wide. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production PREGSF90, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2009, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A.; Misztal, I. Weighting strategies for single-step genomic BLUP: An iterative approach for accurate calculation of GEBV and GWAS. Front. Genet. 2016, 7, 151. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, 183–189. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Faria, C.U.D.; Koury Filho, W.; Magnabosco, C.U.; Albuquerque, L.G.D.; Bezerra, L.A.F.; Lôbo, R.B. Bayesian inference in genetic parameter estimation of visual scores in Nellore beef-cattle. Genet. Mol. Biol. 2009, 32, 753–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertipaglia, T.S.; Carreño, L.O.D.; Machado, C.H.C.; Andrighetto, C.; Fonseca, R. Estimates of Genetic Parameters for Visual Scores and Their Correlation with Production and Reproductive Traits in Brahman Cattle. Rev. Bras. Zootec. 2012, 41, 1407–1411. [Google Scholar] [CrossRef] [Green Version]
- Bonin, M.N.; Ferraz, J.B.S.; Pedrosa, V.B.; Silva, S.L.; Gomes, R.C.; Cucco, D.C.; Santan, M.H.A.; Campos, J.H.A.; Barbosa, V.N.; Castro, F.S.F.; et al. Visual Body-Scores Selection and Its Influence on Body Size and Ultrasound Carcass Traits in Nellore Cattle. J. Anim. Sci. 2015, 93, 5597–5606. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, X.; Deng, D.; Yu, M.; Li, X. Genetic Determinants of Pig Birth Weight Variability. BMC Genet. 2016, 17, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhou, L.; Xie, X.; Wu, Z.; Xiong, X.; Zhang, Z.; Yang, J.; Xiao, S.; Zhou, M.; Ma, J.; et al. Muscle Glycogen Level and Occurrence of Acid Meat in Commercial Hybrid Pigs Are Regulated by Two Low-Frequency Causal Variants with Large Effects and Multiple Common Variants with Small Effects. Genet. Sel. Evol. 2019, 51, 46. [Google Scholar] [CrossRef] [Green Version]
- Twomey, A.J.; Berry, D.P.; Evans, R.D.; Doherty, M.L.; Graham, D.A.; Purfield, D.C. Genome-Wide Association Study of Endo-Parasite Phenotypes Using Imputed Whole-Genome Sequence Data in Dairy and Beef Cattle. Genet. Sel. Evol. 2019, 51, 15. [Google Scholar] [CrossRef] [Green Version]
- Schlegel, G.; Keller, J.; Hirche, F.; Geibler, S.; Schwarz, F.J.; Ringseis, R.; Stangl, G.I.; Eder, K. Expression of Genes Involved in Hepatic Carnitine Synthesis and Uptake in Dairy Cows in the Transition Period and at Different Stages of Lactation. BMC Vet. Res. 2012, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Hoppel, C. The Role of Carnitine in Normal and Altered Fatty Acid Metabolism. Am. J. Kidney Dis. 2003, 41, 4–12. [Google Scholar] [CrossRef]
- Cônsolo, N.R.B.; Silva, J.; Buarque, V.L.M.; Padilla, H.P.; Barbosa, L.C.G.S.; Zawadzki, A.; Colnago, L.A.; Saran Netto, A.; Gerrard, D.E.; Silva, S.L. Selection for Growth and Precocity Alters Muscle Metabolism in Nellore Cattle. Metabolites 2020, 10, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taga, H.; Chilliard, Y.; Meunier, B.; Chambon, C.; Picard, B.; Zingaretti, M.C.; Cinti, S.; Bonnet, M. Cellular and Molecular Large-Scale Features of Fetal Adipose Tissue: Is Bovine Perirenal Adipose Tissue Brown? J. Cell. Physiol. 2012, 227, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.; Hanting, G.; Feng, L.; Ningbo, C.; Yongjie, X.; Jinhang, J.; Fen, L.; Fangru, L.; Man, Z.; Kuilin, S.; et al. Tissue Expression, Association Analysis between Three Novel SNPs of the RXRα Gene and Growth Traits in Chinese Indigenous Cattle. Chin. Sci. Bull. 2013, 58, 2053–2060. [Google Scholar] [CrossRef] [Green Version]
- Janani, C.; Ranjitha Kumari, B.D. PPAR Gamma Gene—A Review. Diabetes Metab. Syndr. Clin. Res. Rev. 2015, 9, 46–50. [Google Scholar] [CrossRef]
- McCabe, M.; Waters, S.; Morris, D.; Kenny, D.; Lynn, D.; Creevey, C. RNA-Seq Analysis of Differential Gene Expression in Liver from Lactating Dairy Cows Divergent in Negative Energy Balance. BMC Genom. 2012, 13, 193. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Li, M.; Prakapenka, D.; Vanraden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Jiang, Z.; Michal, J.J.; Chen, J.; Daniels, T.F.; Kunej, T.; Garcia, M.D.; Gaskins, C.T.; Busboom, J.R.; Alexander, L.J.; Wright, R.W.; et al. Discovery of Novel Genetic Networks Associated with 19 Economically Important Traits in Beef Cattle. Int. J. Biol. Sci. 2009, 5, 528–542. [Google Scholar] [CrossRef] [Green Version]
- Gurgul, A.; Jasielczuk, I.; Szmatoła, T.; Sosin. B., E.; Majewska, A.; Litwińczuk, Z. Divergent Selection Signatures of Phenotypic and Production Traits among Conserved and Commercial Cattle Breeds. Livest. Sci. 2020, 239, 104174. [Google Scholar] [CrossRef]
- Zhang, H.M.; Xia, H.; Jiang, H.; Mao, Y.; Qu, K.; Huang, B.; Gong, Y.; Yang, Z. Ryan. Longissimus Dorsi Muscle Transcriptomic Analysis of Yunling and Chinese Simmental Cattle Differing in Intramuscular Fat Content and Fatty Acid Composition. Genome 2018, 61, 549–558. [Google Scholar] [CrossRef]
- Tisdale, E.J.; Bourne, J.R.; Khosravi-Far, R.; Der, C.J.; Balch, W.E. GTP-Binding Mutants of Rabl and Rab2 Are Potent Inhibitors of Vesicular Transport from the Endoplasmic Reticulum to the Golgi Complex. J. Cell. Biol. 1992, 119, 749–761. [Google Scholar] [CrossRef]
- Gan, Q.F.; Li, Y.R.; Liu, Q.H.; Lund, M.; Su, G.S.; Liang, X.W. Genome-Wide Association Studies for the Concentrations of Insulin, Triiodothyronine, and Thyroxine in Chinese Holstein Cattle. Trop. Anim. Health Prod. 2020, 52, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kim, J.J. Multiple linkage disequilibrium mapping methods to validate additive quantitative trait loci in Korean native cattle (Hanwoo). Asian Australas. J. Anim. Sci. 2015, 28, 926–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seabury, C.M.; Oldeschulte, D.L.; Saatchi, M.; Beever, J.E.; Decker, J.E.; Halley, Y.A.; Bhattarai, E.K.; Molaei, M.; Freetly, H.C.; Hansen, S.L.; et al. Genome-Wide Association Study for Feed Efficiency and Growth Traits in U.S. Beef Cattle. BMC Genom. 2017, 18, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Fortes, M.R.S.; Reverter, A.; Nagaraj, S.H.; Zhang, Y.; Jonsson, N.N.; Barris, W.; Lehnert, S.; Hansen, G.B.B.; Hawken, R.G. A single nucleotide polymorphism–derived regulatory gene network underlying puberty in 2 tropical breeds of beef cattle. J. Anim. Sci. 2011, 89, 1669–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, D.B.S.; Fonseca, L.F.S.; Pinheiro, D.G.; Magalhães, A.F.B.; Muniz, M.M.M.; Ferro, J.A.; BaldI, F.; Chardulo, L.A.L.; Schnabel, R.D.; Taylor, J.F. Spliced Genes in Muscle from Nelore Cattle and Their Association with Carcass and Meat Quality. Sci. Rep. 2020, 10, 1470. [Google Scholar] [CrossRef] [PubMed]
- Vasan, R.S.; Larson, M.G.; Aragam, J.; Wang, T.J.; Mitchell, G.F.; Kathiresan, S.; Newton-Cheh, C.; Vita, J.A.; Keyes, M.J.; O’donnell, C.J.; et al. Genome-Wide Association of Echocardiographic Dimensions, Brachial Artery Endothelial Function and Treadmill Exercise Responses in the Framingham Heart Study. BMC Med. Genet. 2007, 8, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edea, Z.; Kim, K.S. A Whole Genomic Scan to Detect Selection Signatures between Berkshire and Korean Native Pig Breeds. J. Anim. Sci. Technol. 2014, 56, 23. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, M.; Watanabe, T.; Sugimoto, Y. The Molecular Effects of a Polymorphism in the 5′UTR of Solute Carrier Family 44, Member 5 That Is Associated with Birth Weight in Holsteins. PLoS ONE 2012, 7, e41267. [Google Scholar] [CrossRef] [Green Version]
- Fernández, J.C.; Pérez, J.E.; Herrera, N.; Martínez, R.; Bejarano, D.; Rocha, J.F. Genomic Association Study for Age at First Calving and Calving Interval in Romosinuano and Costeño Con Cuernos Cattle. Genet. Mol. Res. 2019, 18, gmr18258. [Google Scholar] [CrossRef]
- Zhang, H.; Shen, L.Y.; Xu, Z.C.; Kramer, L.M.; Yu, J.Q.; Zhang, X.Y.; Na, W.; Yang, L.L.; Cao, Z.P.; Luan, P.; et al. Haplotype-Based Genome-Wide Association Studies for Carcass and Growth Traits in Chicken. Poult. Sci. 2020, 99, 2349–2361. [Google Scholar] [CrossRef]
- Seegar, T.C.M.; Blacklow, S.C. Domain Integration of ADAM Family Proteins: Emerging Themes from Structural Studies. Exp. Biol. Med. 2019, 244, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zan, L.; Zhao, S.; Xin, Y.; Jiao, Y.; Li, K. Molecular Characterization, Expression Pattern, Polymorphism and Association Analysis of Bovine ADAMTSL3 Gene. Mol. Biol. Rep. 2012, 39, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Martínez, R.; Bejarano, D.; Gómez, Y.; Dasoneville, R.; Jiménez, A.; Even, G.; Sölkner, J.; Mészáros, J. Genome-Wide Association Study for Birth, Weaning and Yearling Weight in Colombian Brahman Cattle. Genet. Mol. Biol. 2017, 40, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, S.; Zen, H.; Megens, H.J.; Archibald, A.L.; Haley, C.; Jackson, I.J.; Groenen, M.A.M.; Crooijmans, R.P.M.A.; Ogden, R.; Wiener, P. Signatures of Diversifying Selection in European Pig Breeds. PLoS Genet. 2013, 9, e1003453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, S.H.A.; Khan, R.; Gui, L.; Schreurs, N.M.; Wang, X.; Mei, C.; Yang, X.; Gong, C.; Zan, L. Bioinformatics Analysis and Genetic Polymorphisms in Genomic Region of the Bovine SH2B2 Gene and Their Associations with Molecular Breeding for Body Size Traits in Qinchuan Beef Cattle. Biosci. Rep. 2020, 40, BSR20192113. [Google Scholar] [CrossRef] [Green Version]
- Mu, Q.; Zhu, Z.; Si, Y.; Chen, X.; Duan, G.; Sun, S.; Fang, G.; Zeng, Y.; Yang, N. Overexpression of PID1 Reduces High Density Lipoprotein Level and Functionality in Swine. IUBMB Life 2019, 71, 1946–1951. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.M.; Wang, Y.M.; Zhang, C.M.; Qiu, J.; Shen, Y.H.; Zhu, J.G.; Chen, L.; Xu, G.F.; Zhao, Y.P.; Ji, C.B.; et al. Knockdown of NYGGF4 (PID1) rescues insulin resistance and mitochondrial dysfunction induced by FCCP in 3T3-L1 adipocytes. Mitochondrion 2012, 12, 600–606. [Google Scholar] [CrossRef]
- Zeng, X.Q.; Zeng, X.Q.; Zhang, C.M.; Tong, M.L.; Chi, X.; Li, X.L.; Ji, C.B.; Zhang, R.; Guo, X.R. “Knockdown of NYGGF4 Increases Glucose Transport in C2C12 Mice Skeletal Myocytes by Activation IRS-1/PI3K/AKT Insulin Pathway”. J. Bioenerg. Biomembr. 2012, 44, 351–355. [Google Scholar] [CrossRef]
- Clark, D.L.; Boler, D.D.; Kutzler, L.W.; Jones, K.A.; McKeith, F.K.; Killefer, J.; Carr, T.R.; Dilger, A.C. Muscle Gene Expression Associated with Increased Marbling in Beef Cattle. Anim. Biotechnol. 2011, 22, 51–63. [Google Scholar] [CrossRef]
- Park, J.R.; Jung, J.W.; Seo, M.S.; Kang, S.K.; Lee, Y.S.; Kang, K.S. DNER Modulates Adipogenesis of Human Adipose Tissue-Derived Mesenchymal Stem Cells via Regulation of Cell Proliferation. Cell Prolif. 2010, 43, 19–28. [Google Scholar] [CrossRef]
- Sasaki, S.; Ibi, T.; Ikeda, S.; Sugimoto, Y. A Genome-Wide Association Study Reveals a Quantitative Trait Locus for Age at First Calving in Delta/Notch-like EGF Repeat Containing on Chromosome 2 in Japanese Black Cattle. Anim. Genet. 2014, 45, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Sadkowski, T.; Jank, M.; Zwierzchowski, L.; Oprzadek, J.; Motyl, T. Transcriptomic index of skeletal muscle of beef breeds bulls. J. Physiol. Pharmacol. 2009, 60, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Egerman, M.A.; Glass, D.J. Signaling Pathways Controlling Skeletal Muscle Mass. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Ethier, J.F.; Houde, A.; Lussier, J.G.; Silversides, D.W. Bovine Activin Receptor Type II CDNA: Cloning and Tissue Expression. Mol. Cell. Endocrinol. 1994, 106, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pande, H.O.; Tesfaye, D.; Hoelker, M.; Gebremedhn, S.; Held, E.; Neuhoff, C.; Tholen, E.; Schellander, K.; Wondim, D.S. MicroRNA-424/503 Cluster Members Regulate Bovine Granulosa Cell Proliferation and Cell Cycle Progression by Targeting SMAD7 Gene through Activin Signalling Pathway. J. Ovarian Res. 2018, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Yamakawa, T.; Whitson, R.H.; Li, S.A.; Itakura, K. Modulator Recognition Factor-2 Is Required for Adipogenesis in Mouse Embryo Fibroblasts and 3T3-L1 Cell. Mol. Endocrinol. 2008, 22, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Miao, J.; Xia, J.; Chang, T.; Guangxin, E.; Bao, J.; Jin, S.; Xu, L.; Zhang, L.; Zhu, B.; et al. Identifying Novel Genes for Carcass Traits by Testing G × E Interaction through Genome-Wide Meta-Analysis in Chinese Simmental Beef Cattle. Livest. Sci. 2018, 212, 75–82. [Google Scholar] [CrossRef]
- Rosse, I.C.; Assis, J.G.; Oliveira, F.S.; Leite, L.R.; Araujo, F.; Zerlotini, A.; VolpinI, A.; DominitinI, A.J.; Lopes, B.C.; Arbex, W.A.; et al. Whole Genome Sequencing of Guzerá Cattle Reveals Genetic Variants in Candidate Genes for Production, Disease Resistance, and Heat Tolerance. Mamm. Genome 2017, 28, 66–80. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-Wide Scan of Selection Signatures in Dehong Humped Cattle for Heat Tolerance and Disease Resistance. Anim. Genet. 2020, 51, 292–299. [Google Scholar] [CrossRef]
- Srivastava, S.; Srikanth, K.; Won, S.; Son, J.; Park, J.E.; Park, W.; Chai, H.H.; Lim, D. Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle. Genes 2020, 11, 551. [Google Scholar] [CrossRef]
- Caldas, Y.R.; Renand, G.; Ballester, M.; Saintilan, R.; Rocha, D. Multi-Breed and Multi-Trait Co-Association Analysis of Meat Tenderness and Other Meat Quality Traits in Three French Beef Cattle Breeds. Genet. Sel. Evol. 2016, 48, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, H.; Shimizu, Y.; Tanaka, A.; Takuya, N.; Tabuchi, I.; Oyama, K.; Taniguchi, M.; Mannen, H.; Sasazaki, S. The SNP in the Promoter Region of the Bovine ELOVL5 Gene Influences Economic Traits Including Subcutaneous Fat Thickness. Mol. Biol. Rep. 2013, 40, 3231–3237. [Google Scholar] [CrossRef]
- Huang, W.; Guo, Y.; Du, W.; Zhang, X.; Li, A.; Miao, X. Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 5278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Jiang, P.; Yu, H.; Yang, Y.; Xia, L.; Yang, R.; Fang, X.; Zhao, Z. MiR-21-3p Targets Elovl5 and Regulates Triglyceride Production in Mammary Epithelial Cells of Cow. DNA Cell Biol. 2019, 38, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Tacuma, S.D.; Parales, J.G.; Prom, C.; Chirivi, M.; Laguna, J.; Lock, A.L.; Contreras, G.A. Transcriptomic Profiling of Adipose Tissue Inflammation, Remodeling, and Lipid Metabolism in Periparturient Dairy Cows (Bos Taurus). BMC Genom. 2020, 21, 824. [Google Scholar] [CrossRef]
- Fukunaga, K.; Yamashita, Y.; Yagisawa, T. Copy Number Variations in BOLA-DQA2, BOLA-DQB, and BOLA-DQA5 Show the Genomic Architecture and Haplotype Frequency of Major Histocompatibility Complex Class Genes in Holstein Cows. HLA 2020, 96, 601–609. [Google Scholar] [CrossRef]
- Prajapati, B.M.; Gupta, J.P.; Pandey, D.P.; Parmar, G.A.; Chaudhari, J. Chaudhari. Molecular Markers for Resistance against Infectious Diseases of Economic Importance. Vet. World 2017, 10, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Mota, R.R.; Silva, F.F.; Lopes, P.S.; Tempelman, R.J.; Sollero, B.P.; Aguilar, I.; Cardoso, F.F. Analyses of Reaction Norms Reveal New Chromosome Regions Associated with Tick Resistance in Cattle. Animal 2018, 12, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Prinsen, R.T.M.M.; Rossoni, A.; Gredler, B.; Bieber, A.; Bagnato, A.; Strillacci, M.G. A Genome Wide Association Study between CNVs and Quantitative Traits in Brown Swiss Cattle. Livest. Sci. 2017, 202, 7–12. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Kim, Y.S.; Iqbal, A.; Kim, J.J. A Whole Genome Association Study to Detect Additive and Dominant Single Nucleotide Polymorphisms for Growth and Carcass Traits in Korean Native Cattle, Hanwoo. Asian Australas. J. Anim. Sci. 2017, 30, 8–19. [Google Scholar] [CrossRef]
- Hlongwane, N.L.; Hadebe, K.; Soma, P.; Dzomba, E.F.; Muchadeyi, F.C. Genome Wide Assessment of Genetic Variation and Population Distinctiveness of the Pig Family in South Africa. Front. Genet. 2020, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- DeAndrade, M.P.; Johnson, R.L.; Unger, E.L.; Zhang, L.; Groen, T.V.; Gamble, K.L.; Li, Y. Motor Restlessness, Sleep Disturbances, Thermal Sensory Alterations and Elevated Serum Iron Levels in Btbd9 Mutant Mice. Hum. Mol. Genet. 2012, 21, 3984–3992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.F.; Schnabel, R.D.; Sutovsky, P. Review: Genomics of Bull Fertility. Animal 2018, 12, 172–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.D.; Lee, S.Y.; Jung, E.Y.; Song, S.; Hur, S.J. Quantitative Changes in Peptides Derived from Proteins in Beef Tenderloin (Psoas Major Muscle) and Striploin (Longissimus Lumborum Muscle) during Cold Storage. Food Chem. 2021, 338, 128029. [Google Scholar] [CrossRef]
- Zakharov, V.V.; Mosevitsky, M.I. Oligomeric Structure of Brain Abundant Proteins GAP-43 and BASP1. J. Struct. Biol. 2010, 170, 470–483. [Google Scholar] [CrossRef]
- Guarnieri, S.; Morabito, C.; Paolini, C.; Boncompagni, C.; Pilla, R.; Illic, G.F.; Mariggiò, M.A. Growth Associated Protein 43 Is Expressed in Skeletal Muscle Fibers and Is Localized in Proximity of Mitochondria and Calcium Release Units. PLoS ONE 2013, 8, e53267. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.Y.; Oliveira, H.R.; Schenkel, F.S.; Pedrosa, V.B.; Melka, M.G.; Brito, L.F. Using imputed whole–genome sequence variants to uncover candidate mutations and genes affecting milking speed and temperament in Holstein cattle. J. Dairy Sci. 2020, 103, 10383–10398. [Google Scholar] [CrossRef]
- Zacco, A.; Cooper, V.; Chantler, P.D.; Fisher, S.H.; Horton, H.L.; Levitt, P. Isolation, biochemical characterization and ultrastructural analysis of the limbic system–associated membrane protein (LAMP), a protein expressed by neurons comprising functional neural circuits. J. Neurosc. 1990, 10, 73–90. [Google Scholar] [CrossRef] [Green Version]
- Pimenta, A.F.; Zhukareva, V.; Barbe, M.F.; Reinoso, B.S.; Grimley, C.; Henzel, W.; Fischer, I.; Levitt, P. The Limbic System-Associated Membrane Protein Is an Ig Superfamily Member That Mediates Selective Neuronal Growth and Axon Targeting. Neuron 1995, 15, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Shang, P.; Li, W.; Liu, G.; Zhang, J.; Li, M.; Wu, L.; Wang, K.; Chamba, Y. Identification of LncRNAs and Genes Responsible for Fatness and Fatty Acid Composition Traits between the Tibetan and Yorkshire Pigs. Int. J. Genom. 2019, 19, 5070975. [Google Scholar] [CrossRef]
- Okada, A.; Tomooka, Y. A Role of Sema6A Expressed in Oligodendrocyte Precursor Cells. Neurosci. Lett. 2013, 539, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Carmelo, V.A.O.; Kadarmideen, H.N. Genome-Wide Epistatic Interaction Networks Affecting Feed Efficiency in Duroc and Landrace Pigs. Article 2020, 11, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Wang, C.; Sun, X.; Jiang, S.; Guo, M. Characterization of the Milk Fat Globule Membrane Proteome in Colostrum and Mature Milk of Xinong Saanen Goats. J. Dairy Sci. 2020, 103, 3017–3024. [Google Scholar] [CrossRef]
- Nonneman, D.J.; Shackelford, S.D.; King, D.A.; Wheeler, T.L.; Wiedmann, R.T.; Snelling, W.M.; Rohrer, G.A. Genome-Wide Association of Meat Quality Traits and Tenderness in Swine 1,2. J. Anim. Sci. 2013, 91, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Jemaa, S.B.; Mastrangelo, S.; Lee, S.H.; Lee, J.H.; Boussaha, M. Genome–wide scan for selection signatures reveals novel insights into the adaptive capacity in local North African cattle. Sci. Rep. 2020, 10, 19466. [Google Scholar] [CrossRef] [PubMed]
- Gómez, A.T.; Dier, E.L.B.; Fontanesi, F.; Barrientos, A. HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function. Ceels 2020, 9, 2620. [Google Scholar] [CrossRef]
- Horodyska, J.; Reyer, H.; Wimmers, K.; Trakooljul, N.; Lawlor, P.G.; Hamill, R.M. Transcriptome analysis of adipose tissue from pigs divergent in feed efficiency reveals alteration in gene networks related to adipose growth, lipid metabolism, extracellular matrix, and immune response. Mol. Genet. 2019, 294, 395–408. [Google Scholar] [CrossRef]
- Cardoso, D.F.; Júnior, G.A.F.; Scalez, D.C.B.; Alves, A.A.C.; Magalhães, A.F.B.; Bresolin, T.; Ventura, R.V.; Li, C.; Oliveira, M.C.S.; Porto Neto, L.R.; et al. Uncovering sub-structure and genomic profiles in across-countries subpopulations of Angus cattle. Sci. Rep. 2020, 10, 8770. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, H.; Riker, A.I.; Fodstad, O.; Ledoux, S.P.; Wilson, G.L.; Tan, M. Emerging metabolic targets in cancer therapy. Front. Biosci. 2011, 16, 1844–1860. [Google Scholar] [CrossRef] [Green Version]
- Geng, M.T.; Yao, Y.; Wang, Y.L.; Wu, X.H.; Sun, C.; Li, R.M.; Fu, S.P.; Duan, R.J.; Liu, J.; Hu, X.W.; et al. Structure, expression, and functional analysis of the hexokinase gene family in cassava. Int. J. Mol. Sci. 2017, 18, 1041. [Google Scholar] [CrossRef]
- Verma, P.; Sharma, A.; Sodhi, M.; Thakur, K.; Kataria, R.S.; Niranjan, S.K.; Bharti, V.K.; Kumar, P.; Giri, A.; Kalia, S.; et al. Transcriptome analysis of circulating pbmcs to understand mechanism of high altitude adaptation in native cattle of ladakh region. Sci. Rep. 2018, 8, 7681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gòdia, M.; Castelló, A.; Rocco, M.; Cabrera, B.; Gil, J.E.R.; Balasch, S.; Lewis, C.; Sánchez, A.; Clop, A. Identification of Circular RNAs in Porcine Sperm and Evaluation of Their Relation to Sperm Motility. Sci. Rep. 2020, 10, 7985. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Banerjee, P.; Joshi, J.; Kapoor, P.; Vijh, R.K. Identification of quantitative trait loci for fat percentage in buffaloes. Indian J. Anim. Sci. 2018, 88, 714–723. [Google Scholar] [CrossRef]
- Zappaterra, M.; Zambonelli, P.; Schivazappa, C.; Simoncini, N.; Virgili, R.; Stefanon, B.; Davoli, R. Investigating the Features of PDO Green Hams during Salting: Insights for New Markers and Genomic Regions in Commercial Hybrid Pigs. Animals 2021, 11, 68. [Google Scholar] [CrossRef]
- Ryu, J.; Lee, L. Identification of Contemporary Selection Signatures Using Composite Log Likelihood and Their Associations with Marbling Score in Korean Cattle. Anim. Genet. 2014, 45, 765–770. [Google Scholar] [CrossRef]
- Price, M.P.; Thompson, R.J.; Eshcol, J.O.; Wemmie, J.A.; Benson, C.J. Stomatin Modulates Gating of Acid–sensing Ion Channels. J. Biol. Chem. 2004, 279, 53886–53891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapatsina, L.; Brand, J.; Poole, K.; Daumke, O.; Lewin, G.R. Stomatin-Domain Proteins. Eur. J. Cell Biol. 2012, 91, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Wang, F.; Liu, C.; Xu, X.; Liu, B. TEAD1-Dependent Expression of the FoxO3a Gene in Mouse Skeletal Muscle. BMC Mol. Biol. 2011, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Borel, A.; Eichenberger, D.; Farjanel, J.; Kessler, E.; Gleyzal, C.J.; Hulmes, D.J.S.; Sommer, P.; Font, B. Lysyl oxidase-like protein from bovine aorta. Isolation and maturation to na active form by bone morphogenetic protein-1. J. Biol. Chem. 2001, 276, 48944–48949. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhao, Y.; Gao, J.; Pawlyk, B.; Starcher, B.; Spencer, J.A.; Yanagisawa, H.; Zuo, J.; Li, T. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat. Genet. 2004, 36, 178–182. [Google Scholar] [CrossRef]
- Han, H.; So, H.; Domby, E.; Engle, T. The Relationship of Pulmonary Artery Copper Concentrations and Genes Involved in Copper Homeostasis in Cattle, Swine, and Goats. Asian Australas. J. Anim. Sci. 2012, 25, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Las Heras-Saldana, S.; Clark, S.A.; Duijvesteijn, N.; Gondro, C.; van der Werf, J.H.; Chen, Y. Combining information from genome-wide association and multi-tissue gene expression studies to elucidate factors underlying genetic variation for residual feed intake in Australian Angus cattle. BMC Genom. 2019, 20, 939. [Google Scholar] [CrossRef] [PubMed]




| Variable | N | Mean | ±SD | Minimum | Maximum | σ2a | σ2e | σ2p | h2 (S.E.) |
|---|---|---|---|---|---|---|---|---|---|
| CONF (score) | 20,808 | 3.39 | 0.96 | 1 | 5 | 0.28 | 0.56 | 0.84 | 0.33 + 0.01 |
| PREC (score) | 20,808 | 3.46 | 0.97 | 1 | 5 | 0.32 | 0.53 | 0.85 | 0.37 + 0.01 |
| MUSC (score) | 20,808 | 3.11 | 0.98 | 1 | 5 | 0.33 | 0.52 | 0.85 | 0.38 + 0.02 |
| Chr | SNP Positions | Var (%) | Gene | Gene Name |
|---|---|---|---|---|
| 3 | 3,033,833:3,060,955 | 1.427 | UCK2 | Uridine-cytidine-kinase 2 |
| 3 | 3,164,474:3,192,425 | 1.427 | TMCO1 | Transmembrane and coiled-coil domains 1 Bos taurus (TMCO1), mRNA |
| 3 | 3,212,293:3,249,677 | 1.427 | ALDH9A1 | Aldehyde dehydrogenase 9 family member A1 |
| 3 | 3,310,029:3,310,881 | 1.427 | MGST3 | Microsomal glutathione S-transferase |
| 3 | 3,431,614:3,345,407 | 1.427 | LRRC52 | Leucine-rich repeat containing 52 |
| 3 | 3,470,729:3,536,373 | 1.423 | RXRG | Retinoid X receptor gamma |
| 5 | 90,271,919:90,351,449 | 1.291 | AEBP2 | AE binding protein 2 |
| 5 | 90,610,566:90,837,005 | 1.646 | PLEKHA5 | Pleckstrin homology domain-containing family A member 5 |
| 14 | 23,883,121:24,011,986 | 1.016 | BPNT2 | 3′(2′), 5′-bisphosphate nucleotidase 2 |
| 14 | 24,373,031:24,396,836 | 1.251 | FAM110B | Family with sequence similarity 110 member B |
| 14 | 24,590,812:24,624,435 | 1.368 | UBXN2B | UBX domain-containing protein 2B |
| 14 | 24,651,537:24,675,169 | 2.062 | CYP7A1 | Cholesterol 7α-hydroxylase |
| 14 | 25,079,291:25,258,596 | 1.245 | TOX | Thymocyte selection-associated high mobility group box |
| 14 | 25,866,853:25,887,784 | 2.601 | CA8 | Carbonic anhydrase 8 |
| 14 | 26,217,826:26,253,265 | 3.898 | RAB2A | Member RAS oncogene family |
| 14 | 26,453,389:26,482,710 | 2.677 | CHD7 | Chromodomain-helicase DNA binding protein 7 |
| 14 | 36,011,003:36,195,954 | 1.083 | KCNB2 | Potassium voltage-gated channel subfamily B member 2 |
| 20 | 61,641,222:61,706,531 | 1.995 | CTNND2 | Catenin delta 2 |
| 21 | 24,484,472:24,509,041 | 1.328 | ADAMTSL3 | ADAMTS-like 3 |
| 21 | 24,597,677:24,614,322 | 1.307 | SH3GL3 | SH3 domain containing GRB2-like 3, endophilin A3 |
| 21 | 24,843,672:24,921,366 | 1.307 | HDGFL3 | HDGF-like 3 |
| 21 | 24,933,056:24,947,938 | 1.328 | TM6SF1 | Transmembrane 6 superfamily member 1 |
| 21 | 25,045,675:25,045,726 | 1.452 | BTBD1 | Pleckstrin homology domain containing A5 |
| 21 | 25,194,048:25,229,486 | 1.352 | MORF4L1 | Mortality factor 4-like 1 |
| 27 | 23,270,756:23,916,949 | 1.107 | DLC1 | DLC1 Rho GTPase activating protein |
| 29 | 17,689,798:17,792,365 | 1.197 | GAB2 | GRB2-associated binding protein 2 |
| 29 | 17,825,902:17,853,459 | 1.197 | USP35 | Ubiquitin-specific peptidase 35 |
| Chr | SNP Positions | Var (%) | Gene | Gene Name |
|---|---|---|---|---|
| 2 | 48,421,237:48,808,281 | 1.465 | ACVR2A | Activin A receptor type 2A |
| 2 | 117,076,082:117,094,178 | 3.683 | PID1 | Phosphotyrosine interaction domain containing 1 |
| 2 | 117,441,080:117,623,992 | 3.521 | DNER | Delta/notch-like EGF repeat containing |
| 2 | 117,857,150:117,859,391 | 3.521 | TRIP12 | Thyroid hormone receptor interactor 12 |
| 3 | 21,542,305:21,628,711 | 1.377 | RNF115 | Ring finger protein 115 |
| 3 | 21,653,070:21,722,429 | 1.377 | GPR89A | G-protein coupled receptor 89A |
| 3 | 21,778,330:21,791,863 | 1.377 | GJA8 | Gap junction protein alpha 8 |
| 3 | 21,859,096:21,925,914 | 1.377 | GJA5 | Gap junction protein alpha 5 |
| 3 | 22,006,767:22,098,901 | 1.379 | BCL9 | BCL9 transcription coactivator |
| 8 | 45,324,486:45,403,166 | 1.040 | TJP2 | Tight junction protein 2 |
| 8 | 45,494,923:45,527,302 | 1.041 | FAM189A2 | Family with sequence similarity 189 member A2 |
| 8 | 45,648,545:45,806,337 | 1.041 | APBA1 | Amyloid beta precursor protein binding, family A, member 1 |
| 13 | 1,912,749:2,287,678 | 1.310 | PLCB4 | Phospholipase C beta 4 |
| 13 | 2,522,361:2,553,000 | 1.324 | LAMP5 | Lysosome-associated membrane protein family member 5 |
| 13 | 7,845,572:7,852,824 | 1.142 | FRLT3 | Fibronectin-leucine-rich transmembrane protein 3 |
| 23 | 11,334,019:11,372,125 | 1.192 | CMTR1 | Cap methyltransferase 1 |
| 23 | 11,389,074:11,436,996 | 1.192 | CCDC167 | Coiled-coil domain containing 167 |
| 23 | 11,560,373:11,597,202 | 1.192 | MDGA1 | MAM domain containing glycosylphosphatidylinositol anchor 1 |
| 23 | 11,735,041:12,026,180 | 1.132 | ZFAND3 | AN1-type zinc finger protein 3 |
| 23 | 12,119,175:12,435,186 | 1.132 | BTBD9 | BTB domain containing 9 |
| 23 | 25,088,616:25,099,203 | 5.281 | GSTA2 | Glutathione S-transferase alpha 2 |
| 23 | 25,165,091:25,171,927 | 5.282 | GSTA5 | Glutathione S-transferase alpha 5 |
| 23 | 25,232,601:25,272,669 | 5.281 | CILK1 | Ciliogenesis-associated kinase 1 |
| 23 | 25,438,003:25,474,020 | 5.281 | ELOVL5 | Elongation of very long chain fatty acids 5 |
| 23 | 25,541,686:25,669,376 | 5.281 | BOLA-DQB | Major histocompatibility complex, Class II, DQ beta |
| Chr | SNP Positions | Var (%) | Gene | Gene Name |
|---|---|---|---|---|
| 1 | 60,273,018:60,301,377 | 1.259 | GAP43 | Growth-associated protein 43 |
| 1 | 60,476,799:60,504,874 | 1.236 | LSAMP | Limbic system-associated membrane protein |
| 7 | 36,887,774:37,185,124 | 4.083 | SEMA6A | Semaphorin 6A |
| 7 | 37,857,202:37,909,672 | 2.139 | HIGD2A | HIG1 hypoxia-inducible domain family member 2a |
| 7 | 37,928,274:37,983,978 | 2.139 | FAF2 | Fas-associated factor 2 |
| 7 | 38,116,057:38,126,424 | 2.138 | TSPAN17 | Tetraspanin 17 |
| 7 | 38,306,218:38,331,961 | 1.879 | UNC5A | Unc-5 netrin receptor A |
| 7 | 38,345,801:38,352,118 | 1.878 | HK3 | Hexokinase 3 |
| 7 | 38,385,427:38,410,913 | 1.878 | UIMC1 | Ubiquitin interaction motif containing 1 |
| 9 | 91,134,263:91,182,225 | 2.292 | CNKSR3 | CNKSR 3 family member |
| 9 | 91,496,960:91,538,184 | 2.311 | SCAF8 | SR-related CTD-associated factor 8 |
| 9 | 91,663,611:91,732,039 | 2.324 | TIAM2 | TIAM Rac1 associated GEF 2 |
| 9 | 91,886,978:91,936,136 | 2.323 | CLDN20 | Claudin 20 |
| 16 | 34,020,390:34,166,299 | 1.170 | CEP170 | Centrosomal protein 170 |
| 16 | 34,431,274:34,439,744 | 1.114 | PLD5 | Phospholipase D family member 5 |
| 21 | 34,641,526:34,712,491 | 1.957 | STOML1 | Stomatin-like 1 |
| 21 | 34,664,967:34,672,830 | 1.884 | LOXL1 | Lysyl oxidase-like 1 |
| 21 | 34,785,250:34,813,843 | 1.956 | GZMB | Granzyme B |
| 21 | 35,032,718:35,183,150 | 2.157 | STXBP6 | Syntax-binding protein 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, P.C.; Brito, L.F.; Martins, R.; Pinto, L.F.B.; Silva, M.R.; Pedrosa, V.B. Genome-Wide Association Analysis Reveals Novel Loci Related with Visual Score Traits in Nellore Cattle Raised in Pasture–Based Systems. Animals 2022, 12, 3526. https://doi.org/10.3390/ani12243526
Machado PC, Brito LF, Martins R, Pinto LFB, Silva MR, Pedrosa VB. Genome-Wide Association Analysis Reveals Novel Loci Related with Visual Score Traits in Nellore Cattle Raised in Pasture–Based Systems. Animals. 2022; 12(24):3526. https://doi.org/10.3390/ani12243526
Chicago/Turabian StyleMachado, Pamela C., Luiz F. Brito, Rafaela Martins, Luis Fernando B. Pinto, Marcio R. Silva, and Victor B. Pedrosa. 2022. "Genome-Wide Association Analysis Reveals Novel Loci Related with Visual Score Traits in Nellore Cattle Raised in Pasture–Based Systems" Animals 12, no. 24: 3526. https://doi.org/10.3390/ani12243526
APA StyleMachado, P. C., Brito, L. F., Martins, R., Pinto, L. F. B., Silva, M. R., & Pedrosa, V. B. (2022). Genome-Wide Association Analysis Reveals Novel Loci Related with Visual Score Traits in Nellore Cattle Raised in Pasture–Based Systems. Animals, 12(24), 3526. https://doi.org/10.3390/ani12243526





