Cytogenetic Analysis of the Bimodal Karyotype of the Common European Adder, Vipera berus (Viperidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Mitotic Chromosome Preparation
2.3. Total Preparation of SCs and Immunostaining
2.4. Microscopy
2.5. Image Analysis
3. Results
3.1. Mitotic Metaphase Karyotyping and Karyotypic Formula
3.2. Immunocytochemical Analysis of Meiotic Prophase I Nuclei of V. berus spermatocytes I
3.2.1. Presynaptic Stages
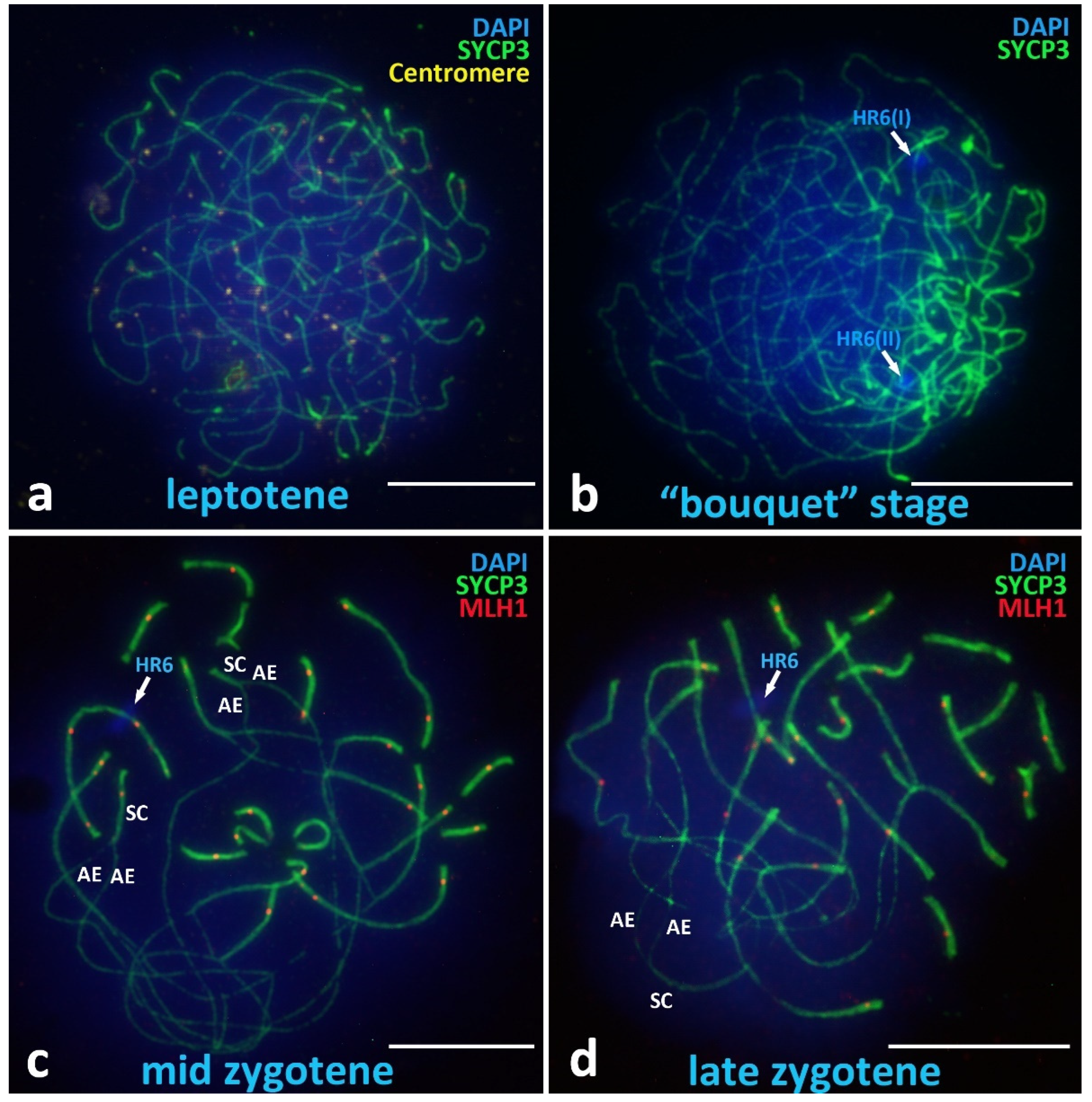
Leptotene
Chromosomal “Bouquet” Stage
Zygotene
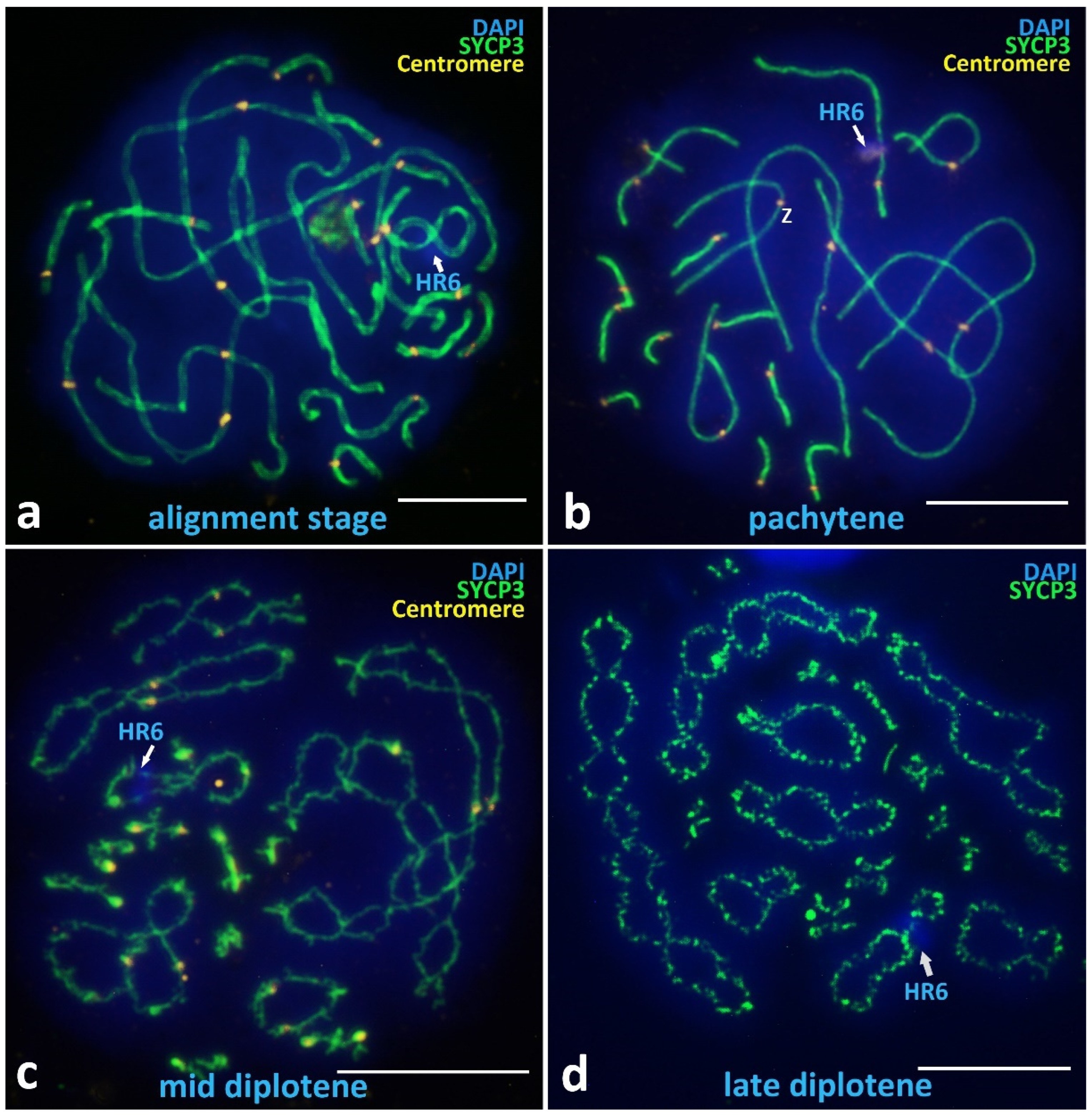
3.2.2. Alignment Stage and Postsynaptic Stages
Alignment Stage
Pachytene
Diplotene
3.2.3. SC-Karyotyping and Meiotic Prophase I Markers Analysis
Crossing-Over Marker, MLH1 Protein
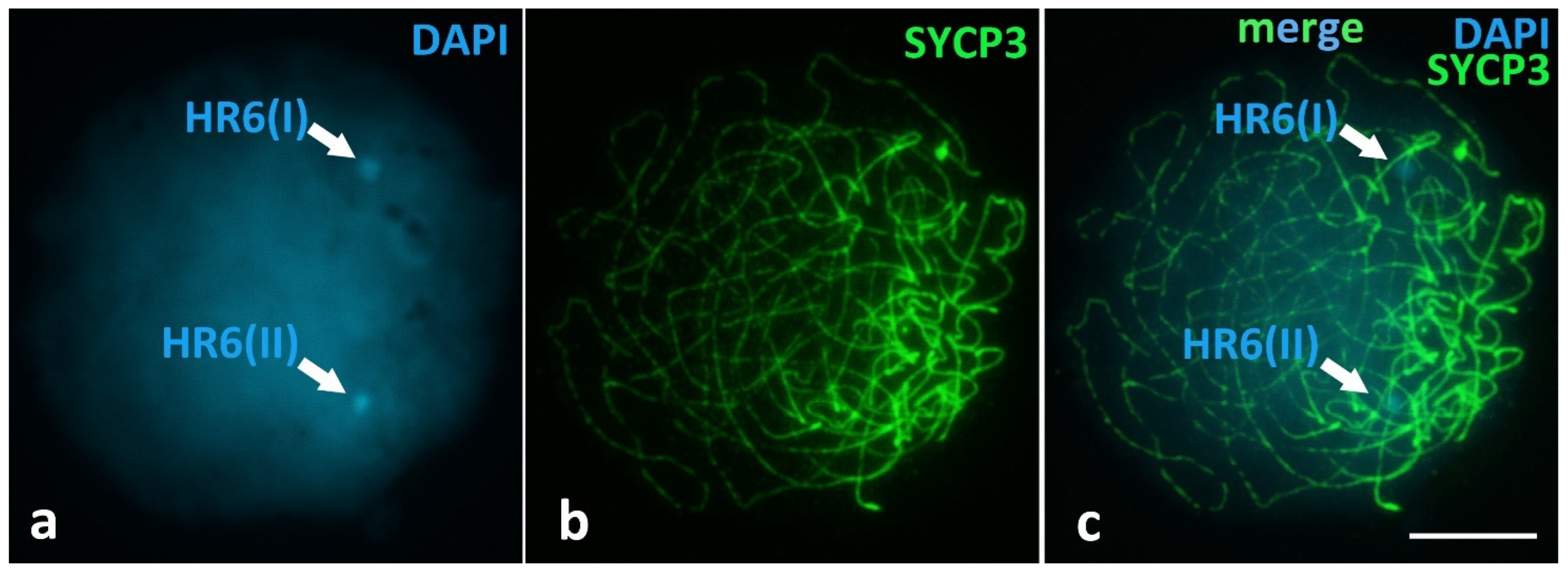
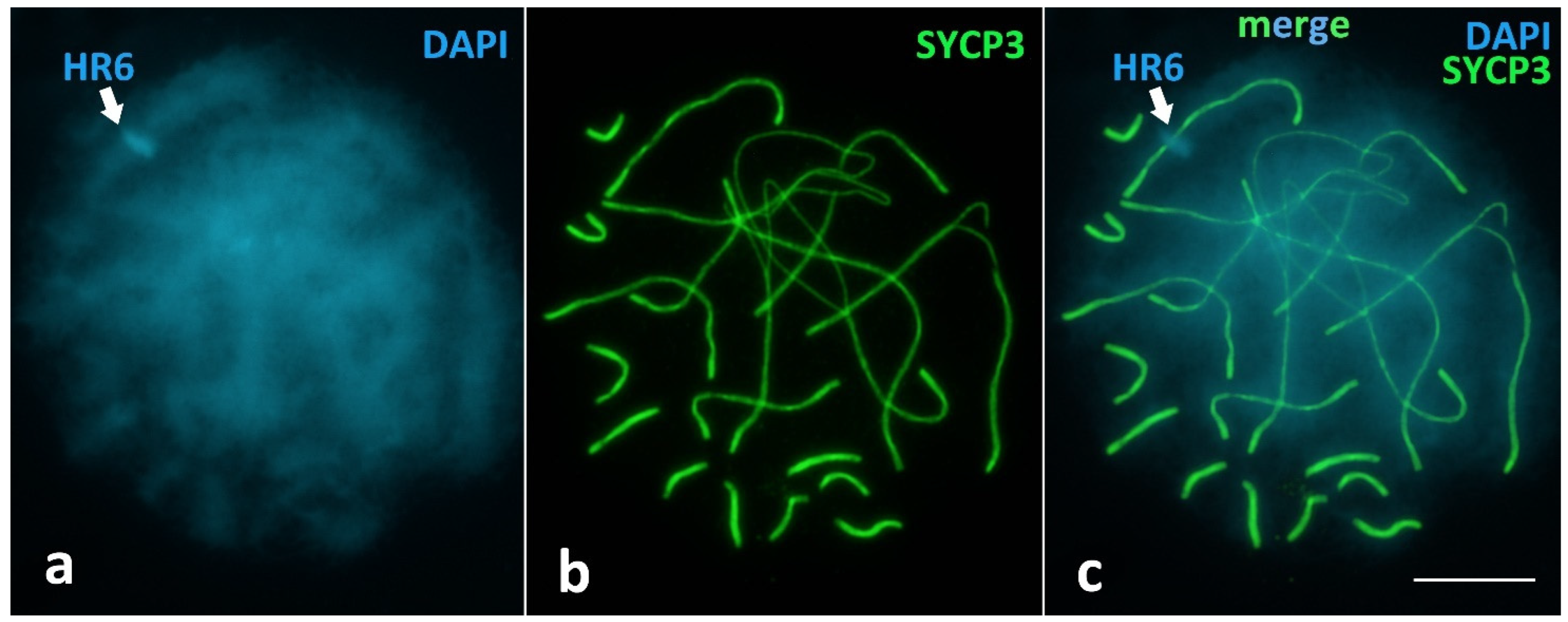
Unique Heterochromatic Chromatin Region (HR6) on the Bivalent 6
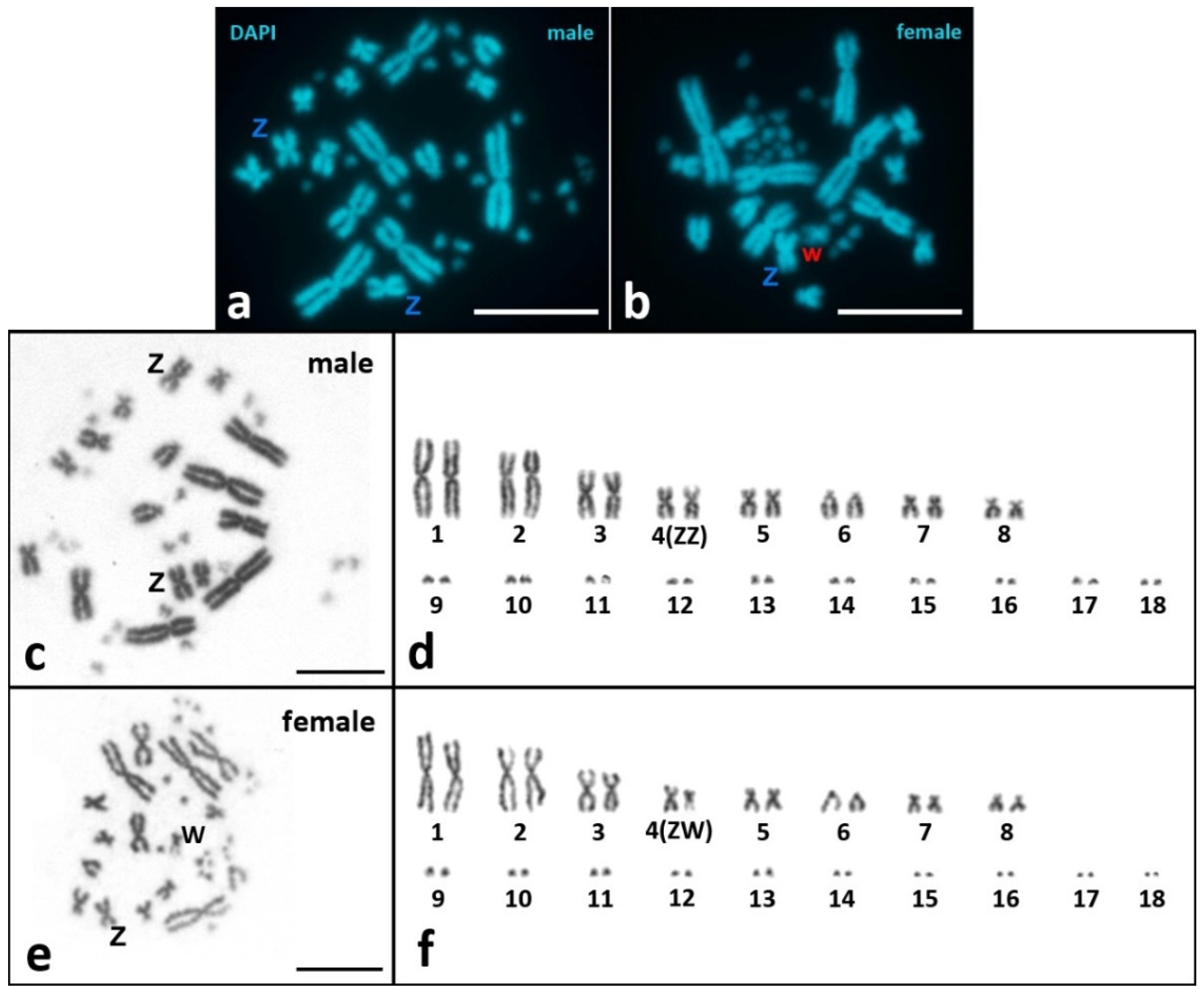
Sex Z Chromosome Identification in the SC-Karyotype
Nucleolar Organizer Region (NOR)
Spermatids
4. Discussion
- Our results in mitotic chromosome karyotyping are in agreement with cytogenetic data obtained previously for V. berus, except for minor differences in the classifications of some macrochromosomes [44]. The karyotype of V. berus described by us consists of 16 macrochromosomes (6m + 8sm + 2st/a) and 20 microchromosomes (2sm + 18st/a), FN = 52. For the first time, we revealed the morphology of microchromosomes in Vipera using high-resolution SC-karyotyping. Karyotypes with 16 macro- and 20 microchromosomes were also described previously in V. ursinii, V. latastei, and V. seoanei [45,81]. On the other hand, V. aspis and V. ammodytes karyotypes with 22 macro- and 20 microchromosomes were reported [45,48,81]. The presence of two variants of karyotypes in the genus Vipera is intriguing, and further studies on both mitotic and meiotic chromosomes are needed to understand the evolution of the karyotypes in the genus.
- Preparations of metaphase plates obtained during the study showed a structured distribution of macro- and microchromosomes: microchromosomes clustered closer to each other, forming a “microchromosome zone”. This was most clearly observed on the weakly and medium-spread mitotic metaphase plates (Figure 1b,e). This partially corroborates the data of other authors, who describe a certain order of arrangement of chromosomes on metaphase plates [67]. Indeed, studies of bimodal karyotypes of birds and reptiles suggest that microchromosomes interact strongly and regularly locate together in somatic nuclei at interphase and during cell division, suggesting their functional coherence [67].
- Synaptonemal complex spread preparations provide detailed visualization of meiotic SC bivalents, which are from three- to five-times longer than mitotic metaphase chromosomes [82]. The V. berus karyotype is a striking example of a bimodal karyotype, combining both very large chromosomes and many microchromosomes. Thus, SC-karyotyping is a logical and very useful method in this regard, as it has allowed us to compare lengths, centromere and NOR positions, and the number of crossing-over sites in microchromosomes.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Phelps, T. Old World Vipers, a Natural History of the Azemiopinae and Viperinae; Edition Chimaira: Frankfurt, Germany, 2010; 558p. [Google Scholar]
- Wüster, W.; Peppin, L.; Pook, C.E.; Walker, D.E. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogen. Evol. 2008, 49, 445–459. [Google Scholar] [CrossRef]
- Szyndlar, Z.; Rage, J.C. Fossil Record of the True Vipers. Biology of the Vipers, Eagle Mountain Publishers 2002, pp. 419–444. Available online: https://scholar.google.com/scholar?hl=ru&as_sdt=0%2C5&q=+Fossil+record+of+the+true+vipers.+Biol.+Vipers+2002%2C+419%E2%80%93444&btnG= (accessed on 10 November 2022).
- Gislen, T.; Kauri, H. Zoogeography of the Swedish amphibians and reptiles. With notes on their growth and ecology. Acta Vertebr. 1959, 1, 197–397. [Google Scholar]
- Archundia, I.G.; de Roodt, A.R.; Ramos-Cerrillo, B.; Chippaux, J.P.; Olguín-Pérez, L.; Alagón, A.; Stock, R.P. Neutralization of Vipera and Macrovipera venoms by two experimental polyvalent antisera: A study of paraspecificity. Toxicon 2011, 57, 1049–1056. [Google Scholar] [CrossRef]
- Gasc, J.P.; Cabela, A.; Crnobrnja-Isailovic, J.; Dolmen, D.; Grossenbacher, K.; Haffner, P.; Lescure, J.; Martens, H.; Martínez-Rica, J.P.; Maurin, H.; et al. Atlas of Amphibians and Reptiles in Europe; Societas Europaea Herpetologica Muséum National d’Histoire Naturelle (IEGB/SPN): Paris, France, 2004; 516p. [Google Scholar]
- Steward, J.W. The Snakes of Europe; Fairleigh Dickinson Univ. Press: Rutherford, NJ, USA, 1971; 238p. [Google Scholar]
- McDiarmid, R.W.; Campbell, J.A.; Touré, T. Snake Species of the World: A Taxonomic and Geographic Reference, Version 1; Herpetologists’ League: Washington, DC, USA, 1999; 511p. [Google Scholar]
- Bannikov, A.G.; Darevskii, I.S.; Ischenko, V.G.; Rustamov, A.K.; Scherbak, N.N. Identification Guide to the Amphibian and Reptile Fauna of the USSR; Moscow “Enlightenment”, USSR: Moscow, Russia, 1977; p. 416. [Google Scholar]
- Zinenko, O.; Sovic, M.; Joger, U.; Gibbs, H.L. Hybrid origin of European Vipers (Vipera magnifica and Vipera orlovi) from the Caucasus determined using genomic scale DNA markers. BMC Evol. Biol. 2016, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Siigur, J.; Siigur, E. Biochemistry and toxicology of proteins and peptides purified from the venom of Vipera berus berus. Toxicon 2022, X, 100131. [Google Scholar] [CrossRef] [PubMed]
- Šmíd, J.; Tolley, K.A. Calibrating the tree of vipers under the fossilized birth-death model. Sci. Rep. 2019, 9, 5510. [Google Scholar] [CrossRef] [PubMed]
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenetics Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Terhivuo, J. Provisional atlas and status of populations for the herpetofauna of Finland in 1980–92. Ann. Zool. Fennici. 1993, 30, 55–69. [Google Scholar]
- Saint Girons, H. Biogeographie et evolution des vipers europeennes. C. R. Soc. Biogeogr. 1980, 56, 146–172. [Google Scholar]
- Nilsson, G.; Andren, C. Vipera berus. In Atlas of Amphibians and Reptiles in Europe; Gasc, J.P., Ed.; Museum National d’Histoire Naturelle: Paris, France; Societas Europaea Herpetologica: Paris, France, 1997; pp. 388–389. [Google Scholar]
- Elmberg, J. Grod- och kräldjurens utbredning i Norrland. Nat. Norr 1995, 14, 57–82. [Google Scholar]
- Andersson, S. Hibernation, habitat and seasonal activity in the adder, Vipera berus, north of the Arctic Circle in Sweden. Amphibia-Reptilia 2003, 24, 449–457. [Google Scholar] [CrossRef]
- Milto, K.D.; Zinenko, O.I. Distribution and morphological variability of Vipera berus in Eastern Europe. Herpetologia Petropolitana. In Proceedings of the 12th Ordinary General Meeting of the Societas Europaea Herpetologica, St. Petersburg, Russia, 12–16 August 2003; Available online: https://www.lacerta.de/AF/Bibliografie/BIB_5285.pdf (accessed on 10 November 2022).
- Bakiev, A.G.; Garanin, V.I.; Gelashvili, D.B.; Gorelov, R.A.; Doronin, I.V.; Zaitseva, O.V.; Zinenko, A.I.; Klyonina, A.A.; Makarova, T.N.; Malenev, A.L.; et al. Vipers (Reptilia: Serpentes: Viperidae: Vipera) of the Volga Basin; Part 1; Kassandra: Togliatti, Russia, 2015; 234p. [Google Scholar]
- Tuniyev, B.S. Rare species of shield-head vipers in the Caucasus. Nature Conservation Research. Zapovednaya Nauka 2016, 1, 11–25. [Google Scholar] [CrossRef]
- Pavlov, A.V. On the results of the study of vipers in the republic of Tatarstan. Mod. Herpetol. 2000, 1, 47–51. [Google Scholar]
- Shiryaev, K.A. New data on reproductive biology of Caucasian species of the genus Vipera. Herpetologia Petropolitana. In Proceedings of the 12th Ordinary General Meeting of the Societas Europaea Herpetologica, St. Petersburg, Russia, 12–16 August 2003; Available online: https://www.lacerta.de/AF/Bibliografie/BIB_5285.pdf (accessed on 10 November 2022).
- Starkov, V.G.; Utkin, Y.N. New data on systematic status of the vipers in Samara Oblast. In Proceedings of the Third Regional Conference of Herpetologists of Volga Basin, Tolyatti, Russia, 5–7 February 2003; pp. 81–82. [Google Scholar]
- Turner, R.K.; Maclean, I.M. Microclimate-driven trends in spring-emergence phenology in a temperate reptile (Vipera berus): Evidence for a potential “climate trap”? Ecol. Evol. 2022, 12, e8623. [Google Scholar] [CrossRef] [PubMed]
- François, D.; Ursenbacher, S.; Boissinot, A.; Ysnel, F.; Lourdais, O. Isolation-by-distance and male-biased dispersal at a fine spatial scale: A study of the common European adder (Vipera berus) in a rural landscape. Conserv. Genet. 2021, 22, 823–837. [Google Scholar] [CrossRef]
- Bauwens, D.; Claus, K. Intermittent reproduction, mortality patterns and lifetime breeding frequency of females in a population of the adder (Vipera berus). PeerJ 2019, 7, e6912. [Google Scholar] [CrossRef]
- Tamagnini, D.; Stephenson, J.; Brown, R.P.; Meloro, C. Geometric morphometric analyses of sexual dimorphism and allometry in two sympatric snakes: Natrix helvetica (Natricidae) and Vipera berus (Viperidae). Zoology 2018, 129, 25–34. [Google Scholar] [CrossRef]
- Nash, D.J.; Griffiths, R.A. Ranging behaviour of adders (Vipera berus) translocated from a development site. Herpetol. J. 2018, 28, 155–159. [Google Scholar]
- Prestt, I. An ecological study of the viper Vipera berus in southern Britain. J. Zool. 1971, 164, 373–418. [Google Scholar] [CrossRef]
- Luiselli, L.; Filippi, E.; Di Lena, E. Ecological relationships between sympatric Vipera aspis and Vipera ursinii in high-altitude habitats of central Italy. J. Herpetol. 2007, 41, 378–384. [Google Scholar] [CrossRef]
- Ursenbacher, S.; Carlsson, M.; Helfer, V.; Tegelström, H.; Fumagalli, L. Phylogeography and Pleistocene refugia of the adder (Vipera berus) as inferred from mitochondrial DNA sequence data. Mol. Ecol. 2006, 15, 3425–3437. [Google Scholar] [CrossRef] [PubMed]
- Starkov, V.G.; Utkin, Y.N. Comparison of venoms of the Vipera genus snakes on the data of cationic exchange chromatography. Actual Probl. Herpetol. Toxinology 2001, 5, 88–89. [Google Scholar]
- Ramazanova, A.S.; Zavada, L.L.; Starkov, V.G.; Kovyazina, I.V.; Subbotina, T.F.; Kostyukhina, E.E.; Dementieva, I.N.; Ovchinnikova, T.V.; Utkin, Y.N. Heterodimeric neurotoxic phospholipases A2—The first proteins from venom of recently established species Vipera nikolskii: Implication of venom composition in viper systematics. Toxicon 2008, 51, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Dyachenko, I.A.; Murashev, A.N.; Andreeva, T.V.; Tsetlin, V.I.; Utkin, Y.N. Analysis of nociceptive effects of neurotoxic phospholipase A2 from Vipera nikolskii venom in mice. J. Venom Res. 2013, 4, 1–4. [Google Scholar] [PubMed]
- Makarova, Y.V.; Kryukova, E.V.; Shelukhina, I.V.; Lebedev, D.S.; Andreeva, T.V.; Ryazantsev, D.Y.; Balandin, S.V.; Ovchinnikova, T.V.; Tsetlin, V.I.; Utkin, Y.N. The first recombinant viper three-finger toxins: Inhibition of muscle and neuronal nicotinic acetylcholine receptors. Dokl. Biochem. Biophys. 2018, 479, 127–130. [Google Scholar] [CrossRef]
- Samel, M.; Vija, H.; Kurvet, I.; Künnis-Beres, K.; Trummal, K.; Subbi, J.; Siigur, J. Interactions of PLA2-s from Vipera lebetina, Vipera berus berus and Naja naja oxiana venom with platelets, bacterial and cancer cells. Toxins 2013, 5, 203–223. [Google Scholar] [CrossRef]
- Cristina, R.T.; Kocsis, R.; Tulcan, C.; Alexa, E.; Boldura, O.M.; Hulea, C.I.; Muselin, F. Protein structure of the venom in nine species of snake: From bio-compounds to possible healing agents. Braz. J. Med. Biol. Res. 2020, 53, e9001. [Google Scholar] [CrossRef]
- Carlsson, M.; Söderberg, L.; Tegelström, H. The genetic structure of adders (Vipera berus) in Fennoscandia: Congruence between different kinds of genetic markers. Mol. Ecol. 2004, 13, 3147–3152. [Google Scholar] [CrossRef]
- Metzger, C.; Ferchaud, A.-L.; Ursenbacher, C.G.S. New polymorphic microsatellite markers of the endangered meadow viper (Vipera ursinii) identified by 454 high-throughput sequencing: When innovation meets conservation. Conserv. Genet. Resour. 2011, 3, 589–592. [Google Scholar] [CrossRef]
- Ursenbacher, S.; Monney, J.C.; Fumagalli, L. Limited genetic diversity and high differentiation among the remnant adder (Vipera berus) populations in the Swiss and French Jura Mountains. Conservation 2009, 10, 303–315. [Google Scholar] [CrossRef]
- Efimov, R.V.; Zav’yalov, E.V.; Velikov, V.A.; Tabachishin, V.G. Genetic divergence of Vipera berus and Vipera nikolskii (Reptilia: Viperidae, Vipera) populations in lower Volga and adjacent territories assessed according to the sequences of cytochrome oxidase III and 12S ribosome RNA genes. Russ. J. Genet. 2008, 44, 240–243. [Google Scholar] [CrossRef]
- Makino, S.; Momma, E. An idiogram study of the chromosomes in some species of reptiles. Cytologia 1949, 15, 96–108. [Google Scholar] [CrossRef]
- Kobel, H.R. Heterochromosomen bei Vipera berus L. (Viperidae, Serpentes). Experientia 1962, 18, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Kobel, H.R. Morphometrische Karyotypanalyse Einiger Schalangen-Arten. Genetica 1967, 38, 1–31. [Google Scholar] [CrossRef]
- Puzachenko, A.Y.; Baklushinskaya, I.Y.; Lyapunova, E.A.; Vlasov, A.A. Karyotype of Vipera ursini (Reptilia, Viperidae) from Streletskaya steppe. (In Russian, English summary). Vestn. Zool. 1997, 31, 81–82. [Google Scholar]
- Zavialov, E.V.; Kaybeleva, E.I.; Tabachishin, V.G. A comparative karyological characteristics of Forest-Steppe Viper (Vipera (Pelias) nikolskii) from the small river flood-lands of the Volga and Don basins. Curr. Stud. Herpetol. 2006, 5/6, 100–103. [Google Scholar]
- Aprea, G.; Odierna, G.; Gentilli, A.; Zuffi, M. The karyology of Vipera aspis, V. atra, V. hugyi, and Cerastes vipera. Amphibia-Reptilia 2006, 27, 113–119. [Google Scholar]
- Zinenko, O. New data about hybridization between Vipera nikolskii (Vedmederya, Grubant et Rudaeva, 1986) and Vipera berus berus (Linnaeus, 1758) and their contact zones in Ukraine. Mertensiella 2004, 15, 17–28. [Google Scholar]
- Martínez-Freiría, F.; Lizana, M.; do Amaral, J.P.; Brito, J. Spatial and temporal segregation allows coexistence in a hybrid zone among two Mediterranean vipers (Vipera aspis and V. latastei). Amphibia-Reptilia 2010, 31, 195–212. [Google Scholar] [CrossRef]
- Pavlov, A.V.; Zinenko, O.I.; Joger, U.; Stümpel, N.; Petrova, I.V.; Malenyov, A.L.; Zaitseva, O.V.; Shurshina, I.V.; Bakiev, A.G. Natural hybridization of the Eastern Steppe Viper Vipera renardi and the Common Adder V. berus. Proc. Samara Sci. Cent. Russ. Acad. Sci. 2011, 13, 172–178. [Google Scholar]
- Czirják, G.Á.; Köbölkuti, L.B.; Tenk, M.; Szakács, A.; Kelemen, A.; Spînu, M. Hemorrhagic stomatitis in a natural hybrid of Vipera ammodytes × Vipera berus due to inappropriate substrate in terrarium. J. Vet. Med. Sci. 2015, 77, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Guiller, G.; Lourdais, O.; Ursenbacher, S. Hybridization between a Euro-Siberian (Vipera berus) and a Para-Mediterranean viper (V. aspis) at their contact zone in western France. J. Zool. 2016, 302, 138–147. [Google Scholar] [CrossRef]
- Zinenko, O.I. First generation hybrids between the Nikolsky’s adder, Vipera nikolskii, and the common adder (Reptilia, Serpentes, Viperidae). Vestn. Zool. Kiev 2003, 37, 101–104. [Google Scholar]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Seki, R.; Nishida, C.; Matsuda, Y. Molecular cloning and characterization of satellite DNA sequences from constitutive heterochromatin of the habu snake (Protobothrops flavoviridis, Viperidae) and the Burmese python (Python bivittatus, Pythonidae). Chromosoma 2015, 124, 529–539. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerful facilitators of chromosomal rearrangements for diversity and evolution. Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Rodionov, A.V. Micro versus macro: A review of structure and functions of avian micro-and macrochromosomes. Russ. J. Genet. 1996, 32, 517–527. [Google Scholar]
- Pigozzi, M.I. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet. Genome Res. 2001, 95, 129–133. [Google Scholar] [CrossRef]
- Wilcox, J.J.; Arca-Ruibal, B.; Samour, J.; Mateuta, V.; Idaghdour, Y.; Boissinot, S. Linked-Read Sequencing of Eight Falcons Reveals a Unique Genomic Architecture in Flux. Genome Biol. Evol. 2022, 14, evac090. [Google Scholar] [CrossRef]
- Morescalchi, A.; Odierna, G.; Olmo, E. Karyological relationships between the Cryptobranchid salamanders. Experientia 1977, 33, 1579–1581. [Google Scholar] [CrossRef]
- Stingo, V.; Rocco, L. Chondrichthyan cytogenetics: A comparison with teleosteans. J. Mol. Evol. 1991, 33, 76–82. [Google Scholar] [CrossRef]
- Rock, J.; Eldridge, M.; Champion, A.; Johnston, P.; Joss, J. Karyotype and nuclear DNA content of the Australian lungfish, Neoceratodus forsteri (Ceratodidae: Dipnoi). Cytogenet. Genome Res. 1996, 73, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Menezes, R.S.; Gazoni, T.; Costa, M.A. Cytogenetics of warrior wasps (Vespidae: Synoeca) reveals intense evolutionary dynamics of ribosomal DNA clusters and an unprecedented number of microchromosomes in Hymenoptera. Biol. J. Linn. Soc. 2019, 126, 925–935. [Google Scholar] [CrossRef]
- Srikulnath, K.; Ahmad, S.F.; Singchat, W.; Panthum, T. Why do some vertebrates have microchromosomes? Cells 2021, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Schield, D.R.; Card, D.C.; Hales, N.R.; Perry, B.W.; Pasquesi, G.M.; Blackmon, H.; Adams, R.H.; Corbin, A.B.; Smith, C.F.; Ramesh, B.; et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019, 29, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, C.; Xu, N.; Bian, P.; Tian, X.; Wang, X.; Yang, N. De Novo Assembly of 20 Chicken Genomes Reveals the Undetectable Phenomenon for Thousands of Core Genes on Microchromosomes and Subtelomeric Regions. Mol. Biol. Evol. 2022, 39, msac066. [Google Scholar] [CrossRef]
- Waters, P.D.; Patel, H.R.; Ruiz-Herrera, A.; Álvarez-González, L.; Lister, N.C.; Simakov, O.; Ezaz, T.; Kaur, P.; Frere, C.; Grützner, F.; et al. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2112494118. [Google Scholar] [CrossRef]
- Olmo, E. Rate of chromosome changes and speciation in reptiles. Genetica 2005, 125, 185–203. [Google Scholar] [CrossRef]
- Spangenberg, V. FISH—And the Characterization of Synaptonemal Complex. In Cytogenetics and Molecular Cytogenetics; CRC Press: Boca Raton, FL, USA, 2022; pp. 297–305. [Google Scholar]
- Spangenberg, V.; Arakelyan, M.; Galoyan, E.; Matveevsky, S.; Petrosyan, R.; Bogdanov, Y.; Danielyan, F.; Kolomiets, O. Reticulate evolution of the rock lizards: Meiotic chromosome dynamics and spermatogenesis in diploid and triploid males of the genus Darevskia. Genes 2017, 8, 149. [Google Scholar] [CrossRef]
- Spangenberg, V.; Arakelyan, M.; Galoyan, E.; Pankin, M.; Petrosyan, R.; Stepanyan, I.; Grishaeva, T.; Danielyan, F.; Kolomiets, O. Extraordinary centromeres: Differences in the meiotic chromosomes of two rock lizards species Darevskia portschinskii and Darevskia raddei. PeerJ 2019, 7, e6360. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- McDermott, A. Human male meiosis: Chromosome behavior at pre-meiotic and meiotic stages of spermatogenesis. Canadian J. Genet. Cytol. 1971, 13, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999, 33, 603. [Google Scholar] [CrossRef] [PubMed]
- Gribbins, K.M. Reptilian spermatogenesis: A histological and ultrastructural perspective. Spermatogenesis 2011, 1, 250–269. [Google Scholar] [CrossRef]
- Viana, P.F.; Feldberg, E.; de Bello Cioffi, M.; de Carvalho, V.T.; Menezes, S.; Vogt, R.C.; Liehr, T.; Ezaz, T. The Amazonian Red Side-Necked Turtle Rhinemys rufipes (Spix, 1824) (Testudines, Chelidae) Has a GSD Sex-Determining Mechanism with an Ancient XY Sex Microchromosome System. Cells 2020, 9, 2088. [Google Scholar] [CrossRef]
- Viana, P.F.; Ezaz, T.; de Bello Cioffi, M.; Liehr, T.; Al-Rikabi Goll, L.G.; Rocha, A.M.; Feldberg, E. Landscape of snake’sex chromosomes evolution spanning 85 MYR reveals ancestry of sequences despite distinct evolutionary trajectories. Sci. Rep. 2020, 10, 12499. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Iannucci, A.; Altmanová, M.; Ciofi, C.; Ferguson-Smith, M.; Pereira, J.C.; Rehák, I.; Stanyon, R.; Velenský, P.; Rovatsos, M.; Kratochvíl, L.; et al. Isolating chromosomes of the Komodo dragon: New tools for comparative mapping and sequence assembly. Cytogenet. Genome Res. 2019, 157, 123–131. [Google Scholar] [CrossRef]
- Iannucci, A.; Makunin, A.I.; Lisachov, A.P.; Ciofi, C.; Stanyon, R.; Svartman, M.; Trifonov, V.A. Bridging the gap between vertebrate cytogenetics and genomics with Single-Chromosome Sequencing (ChromSeq). Genes 2021, 12, 124. [Google Scholar] [CrossRef]
- Saint Girons, R.; Fons, R. Un cas de mélanisme chez Vipera aspis dans les Pyrénées. Vie Milieu 1977, 27, 145–146. [Google Scholar]
- Kalikinskaya, E.I.; Kolomiets, O.L.; Shevchenko, V.A.; Bogdanov, Y.F. Chromosome aberrations in F1 from irradiated male mice studied by their synaptonemal complexes. Mutat. Res. Lett. 1986, 174, 59–65. [Google Scholar] [CrossRef]
- Lisachov, A.P.; Solovyeva, E.N. The toad-headed agamas (Phrynocephalus) are champions in crossing over rate. In Molecular cytogenetics; Biomed Central Ltd.: London, UK, 2017; Volume 10. [Google Scholar]
- Axelsson, E.; Webster, M.T.; Smith, N.G.; Burt, D.W.; Ellegren, H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005, 15, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Cobror, O.; Olmo, E.; Odierna, G.; Angelini, F.; Ciarcia, G. Cyclic variation of chiasma frequency and distribution in Podarcis sicula (Reptilia: Lacertidae). Genetica 1986, 71, 31–37. [Google Scholar] [CrossRef]
- Spangenberg, V.; Arakelyan, M.; Galoyan, E.; Martirosyan, I.; Bogomazova, A.; Martynova, E.; de Bello Cioffi, M.; Liehr, T.; Al-Rikabi, A.; Osipov, F.; et al. Meiotic synapsis of homeologous chromosomes and mismatch repair protein detection in the parthenogenetic rock lizard Darevskia unisexualis. Mol. Reprod. Dev. 2021, 88, 119–127. [Google Scholar] [CrossRef]
- Oguiura, N.; Ferrarezzi, H.; Batistic, R.F. Cytogenetics and Molecular Data in Snakes: A Phylogenetic Approach. Cytogenet. Genome Res. 2009, 127, 128–142. [Google Scholar] [CrossRef]
- Trajtengertz, I.; Beçak, M.L.; Ruiz, I.R. Ribosomal cistrons in Bothrops neuwiedi (Serpentes) subspecies from Brazil. Genome 1995, 38, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.A.; Hamilton, M.J.; Sites, J.W., Jr.; Baker, R.J. Location of ribosomal DNA in chromosomes of squamate reptiles: Systematic and evolutionary implications. Herpetologica 1991, 47, 271–280. [Google Scholar]
- Mezzasalma, M.; Odierna, G. Sex chromosome diversification in the smooth snake Coronella austriaca (Reptilia, Serpentes). Acta Herpetol. 2021, 16, 37–44. [Google Scholar] [CrossRef]
- Cole, C.J.; Hardy, L.M. Karyotypes of six species of colubrid snakes from the Western Hemisphere, and the 140-million-year-old ancestral karyotype of Serpentes. Am. Mus. Novit. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Panthum, T.; Srikulnath, K. Impact of repetitive DNA elements on snake genome biology and evolution. Cells 2021, 10, 1707. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spangenberg, V.; Redekop, I.; Simanovsky, S.A.; Kolomiets, O. Cytogenetic Analysis of the Bimodal Karyotype of the Common European Adder, Vipera berus (Viperidae). Animals 2022, 12, 3563. https://doi.org/10.3390/ani12243563
Spangenberg V, Redekop I, Simanovsky SA, Kolomiets O. Cytogenetic Analysis of the Bimodal Karyotype of the Common European Adder, Vipera berus (Viperidae). Animals. 2022; 12(24):3563. https://doi.org/10.3390/ani12243563
Chicago/Turabian StyleSpangenberg, Victor, Ilya Redekop, Sergey A. Simanovsky, and Oxana Kolomiets. 2022. "Cytogenetic Analysis of the Bimodal Karyotype of the Common European Adder, Vipera berus (Viperidae)" Animals 12, no. 24: 3563. https://doi.org/10.3390/ani12243563
APA StyleSpangenberg, V., Redekop, I., Simanovsky, S. A., & Kolomiets, O. (2022). Cytogenetic Analysis of the Bimodal Karyotype of the Common European Adder, Vipera berus (Viperidae). Animals, 12(24), 3563. https://doi.org/10.3390/ani12243563




