Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Orthologous Gene Set
2.2. Phylogenetic Analysis and Divergence Time Estimation
2.3. Gene Family Expansion and Contraction Analysis
2.4. Rapidly Evolving and Positive Selection Genes Analysis
3. Results
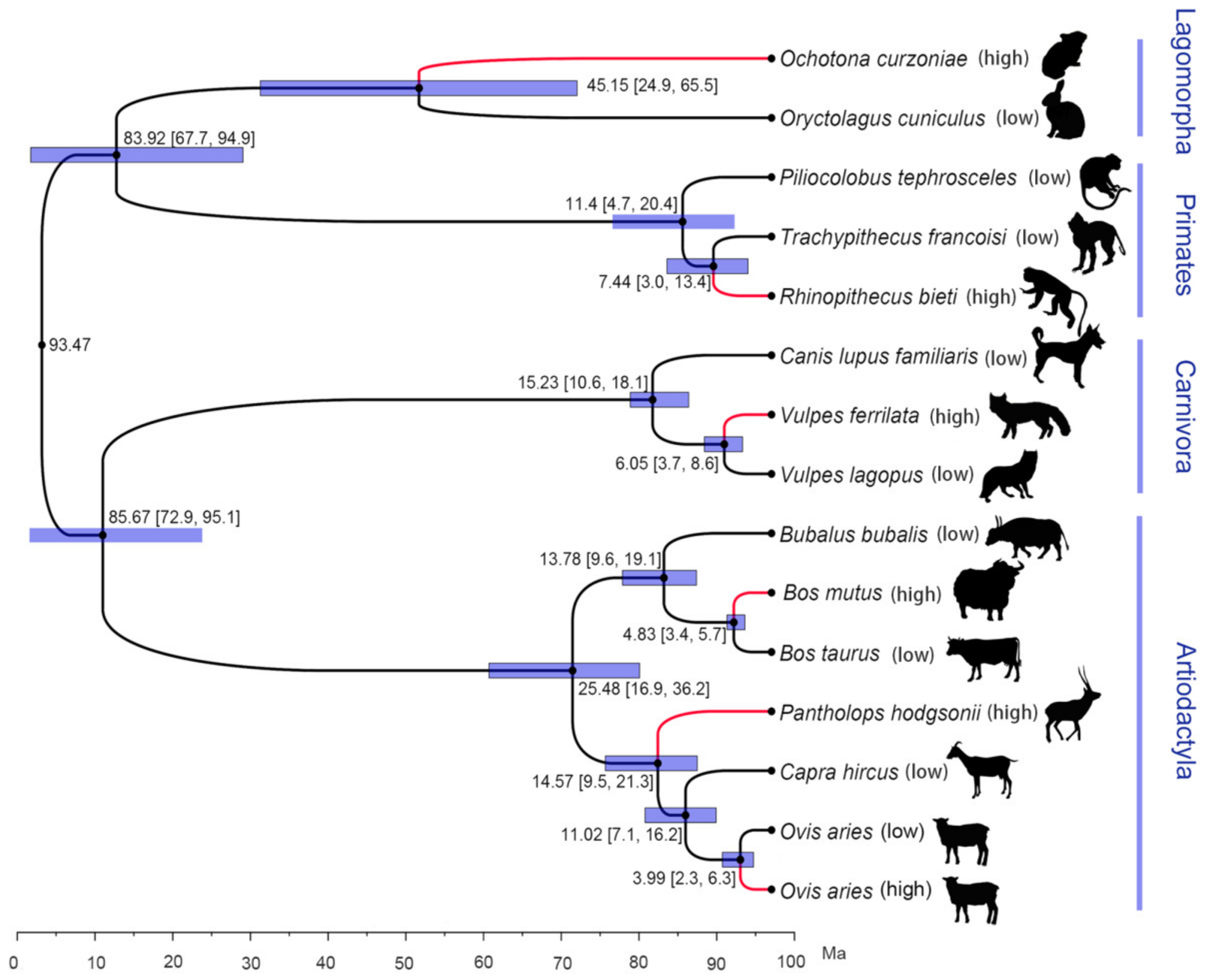
3.1. Identification of Orthologous Gene Set and Phylogenetic Analysis
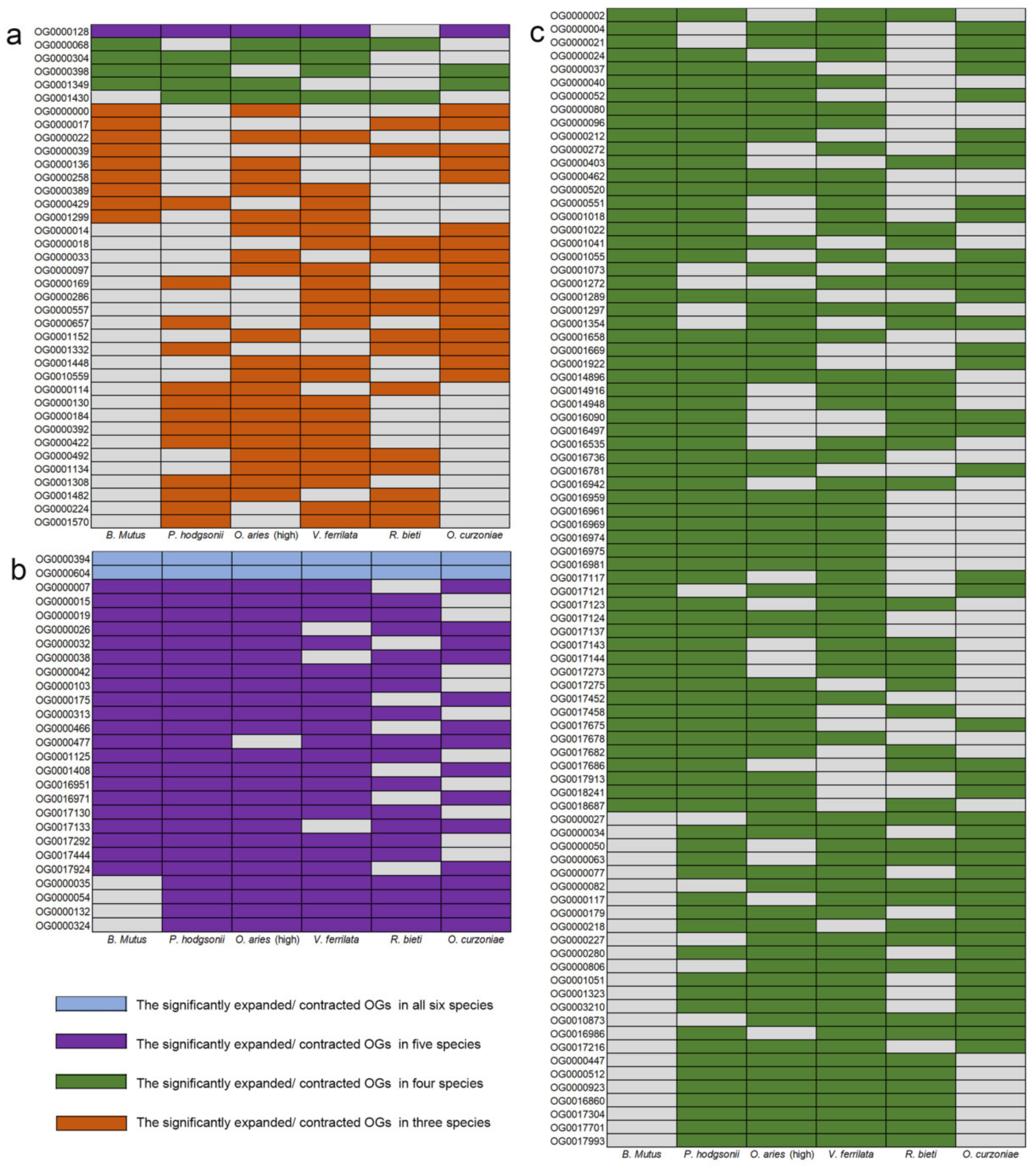
3.2. Gene Family Expansion and Contraction Analysis
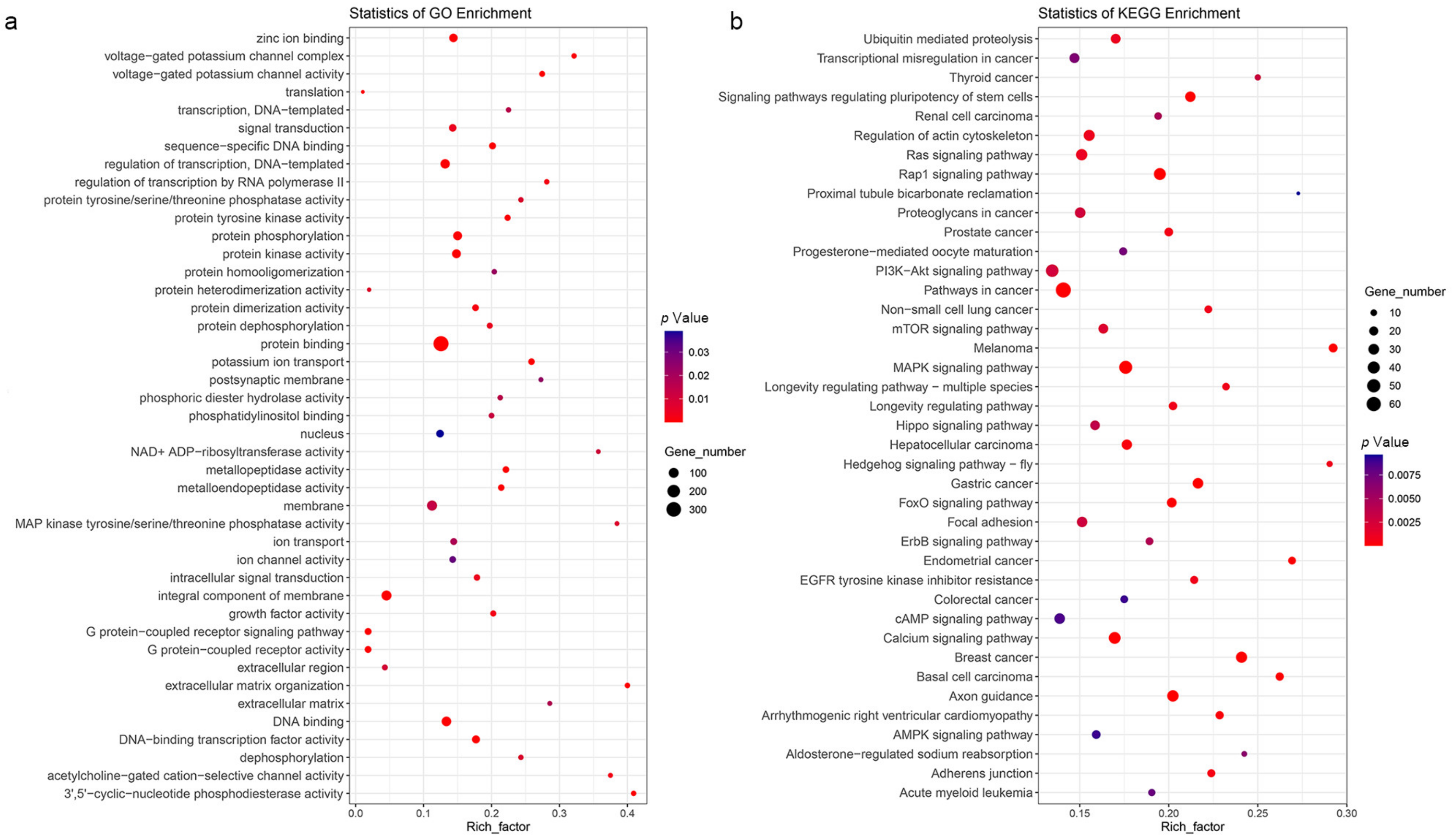
3.3. Rapidly Evolving and Positive Selection Genes
4. Discussion
4.1. The Convergent Adaption Pattern of Gene Family Expansion and Contraction
4.2. The Convergent Adaption Pattern of REGs and PSGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, Q.; Qian, X.; Lang, Y.; Luo, Y.; Xu, J.; Pan, S.; Hui, Y.; Gou, C.; Cai, Y.; Hao, M.; et al. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biol. 2013, 14, R29. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Li, S.; Yang, Y.; Tan, J.; Lou, H.; Jin, W.; Yang, L.; Pan, X.; Wang, J.; Shen, Y.; et al. A Genome-Wide Search for Signals of High-Altitude Adaptation in Tibetans. Mol. Biol. Evol. 2011, 28, 1003–1011. [Google Scholar] [CrossRef] [Green Version]
- Ge, R.L.; Cai, Q.L.; Shen, Y.Y.; San, A.; Ma, L.; Zhang, Y.; Yi, X.; Chen, Y.; Yang, L.F.; Huang, Y.; et al. Draft genome sequence of the Tibetan antelope. Nat. Commun. 2013, 4, 1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, T.S.; Wei, Q.G.; Wang, L.D.; Zhou, S.Y.; Shi, L.P.; Dong, Y.H.; Dou, H.S.; Sha, W.L.; Ga, T.; Zhang, H.H. High-quality chromosome-level genome assembly of Tibetan fox (Vulpes ferrilata). Zool. Res. 2022, 43, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Gao, Y.D.; Xie, L.; Deng, C.; Shi, P.; Guan, M.L.; Huang, S.; Ren, J.L.; Wu, D.D.; Ding, L.; et al. Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc. Natl. Acad. Sci. USA 2018, 115, 8406–8411. [Google Scholar] [CrossRef] [Green Version]
- Bai, L.; Liu, B.N.; Ji, C.M.; Zhao, S.H.; Liu, S. Hypoxic and Cold Adaptation Insights from the Himalayan Marmot Genome. iScience 2019, 11, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, D.; Liu, G.J.; Hou, L.; Gui, W.Y.; Chen, B.; Kang, L. Genetic variation in PTPN1 contributes to metabolic adaptation to high-altitude hypoxia in Tibetan migratory locusts. Nat. Commun. 2018, 9, 4991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabora, L. Convergent evolution. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; Volume 2, pp. 178–180. [Google Scholar]
- Zhang, J.; Kumar, S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 1997, 14, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.Q.; Chu, S.Z.; Shen, Q.S.; Jiang, L.P.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Houliez, E.; Lefebvre, S.; Dessier, A.; Huret, M.; Marquis, E.; Bréret, M.; Dupuy, C. Spatio-temporal drivers of microphytoplankton community in the Bay of Biscay: Do species ecological niches matter? Prog. Oceanogr. 2021, 194, 102558. [Google Scholar] [CrossRef]
- Stern, D.L. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013, 14, 751–764. [Google Scholar] [CrossRef]
- Hu, Y.B.; Wu, Q.; Ma, S.; Ma, T.X.; Shan, L.; Wang, X.; Nie, Y.G.; Ning, Z.M.; Yan, L.; Xiu, Y.F.; et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. USA 2017, 114, 1081–1086. [Google Scholar] [CrossRef] [Green Version]
- Alfieri, F.; Botton-Divet, L.; Nyakatura, J.A.; Amson, E. Integrative Approach Uncovers New Patterns of Ecomorphological Convergence in Slow Arboreal Xenarthrans. J. Mamm. Evol. 2022, 29, 283–312. [Google Scholar] [CrossRef]
- Foote, A.D.; Liu, Y.; Thomas, G.W.C.; Vinař, T.; Alföldi, J.; Deng, J.; Dugan, S.; van Elk, C.E.; Hunter, M.E.; Joshi, V.; et al. Convergent evolution of the genomes of marine mammals. Nat. Genet. 2015, 47, 272–275. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.Y.; Liang, L.; Li, G.S.; Murphy, R.W.; Zhang, Y.P. Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 2012, 8, e1002788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Jin, H.; Fu, J. Molecular convergent and parallel evolution among four high-elevation anuran species from the Tibetan region. BMC Genom. 2020, 21, 839. [Google Scholar] [CrossRef]
- Wang, G.D.; Fan, R.X.; Zhai, W.W.; Liu, F.; Wang, L.; Zhong, L.; Wu, H.; Yang, H.C.; Wu, S.F.; Zhu, C.L.; et al. Genetic Convergence in the Adaptation of Dogs and Humans to the High-Altitude Environment of the Tibetan Plateau. Genome Biol. Evol. 2014, 6, 2122–2128. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Yang, C.; Shen, Q.; Pan, S.; Liu, Z.; Zhang, T.; Zhou, X.; Lei, M.; Chen, P.; Yang, H.; et al. A single mutation underlying phenotypic convergence for hypoxia adaptation on the Qinghai-Tibetan Plateau. Cell Res. 2021, 31, 1032–1035. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Z.; Gu, Y.R.; Zhou, X.M.; Jin, L.; Guan, J.Q.; Liu, R.; Li, J.; Long, K.R.; Tian, S.L.; Che, T.D.; et al. Comparative transcriptomics of 5 high-altitude vertebrates and their low-altitude relatives. GigaScience 2017, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, G.; Ruan, J.; Chen, Y.; Yang, C.; Cao, X.; Wu, H.; Liu, Y.; Du, Z.; Wang, X.; et al. Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 2016, 48, 947–952. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.X.; Wang, J.; Li, Y.F.; Zhou, C.R.; Meng, G.L.; Li, F.J.; Lan, Y.; Price, M.; Podsiadlowski, L.; Yu, Y.; et al. Genomics and morphometrics reveal the adaptive evolution of pikas. Zool. Res. 2022, 43, 813–826. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, X.; Ding, W.; Tian, L.; Zhao, H.; Lu, X. Potential short-term effects of yak and Tibetan sheep dung on greenhouse gas emissions in two alpine grassland soils under laboratory conditions. Biol. Fertil. Soils. 2013, 49, 1215–1226. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar]
- Hassanin, A.; An, J.; Ropiquet, A.; Nguyen, T.T.; Couloux, A. Combining multiple autosomal introns for studying shallow phylogeny and taxonomy of Laurasiatherian mammals: Application to the tribe Bovini (Cetartiodactyla, Bovidae). Mol. Phylogenet. Evol. 2013, 66, 766–775. [Google Scholar] [CrossRef]
- Peng, Y.D.; Li, H.; Liu, Z.Z.; Zhang, C.S.; Li, K.Q.; Gong, Y.F.; Geng, L.Y.; Su, J.J.; Guan, X.M.; Liu, L.; et al. Chromosome-level genome assembly of the Arctic fox (Vulpes lagopus) using PacBio sequencing and Hi-C technology. Mol. Ecol. Resour. 2021, 21, 2093–2108. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.W.; Koehler, A.M.; Crouch, J.A.; Cubeta, M.A.; LeBlanc, N.R. Comparative genomic analysis reveals contraction of gene families with putative roles in pathogenesis in the fungal boxwood pathogens Calonectria henricotiae and C. pseudonaviculata. BMC Ecol. Evol. 2022, 22, 79. [Google Scholar] [CrossRef] [PubMed]
- Demuth, J.P.; Hahn, M.W. The life and death of gene families. Bioessays 2009, 31, 29–39. [Google Scholar] [CrossRef]
- Szűcs, M.; Vahsen, M.L.; Melbourne, B.A.; Hoover, C.; Weiss-Lehman, C.; Hufbauer, R.A. Rapid adaptive evolution in novel environments acts as an architect of population range expansion. Proc. Natl. Acad. Sci. USA 2017, 114, 13501–13506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, L.; Nery, M.F. Expansions and contractions in gene families of independently-evolved blood-feeding insects. BMC Evol. Biol. 2020, 20, 87. [Google Scholar] [CrossRef]
- Albalat, R.; Cañestro, C. Evolution by gene loss. Nat. Rev. Genet. 2016, 17, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Mage, R.G.; Pinheiro, A.; de Matos, A.L.; Esteves, P.J. The Immune System of Lagomorphs. Encycl. Immunobiol. 2016, 1, 515–525. [Google Scholar]
- Hottes, A.K.; Freddolino, P.L.; Khare, A.; Donnell, Z.N.; Liu, J.C.; Tavazoie, S. Bacterial Adaptation through Loss of Function. PLoS Genet. 2013, 7, e1003617. [Google Scholar] [CrossRef]
- Kvitek, D.J.; Sherlock, G. Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment. PLoS Genet. 2013, 11, e1003972. [Google Scholar] [CrossRef] [Green Version]
- Cooper, V.S.; Schneider, D.; Blot, M.; Lenski, R.E. Mechanisms Causing Rapid and Parallel Losses of Ribose Catabolism in Evolving Populations of Escherichia coli B. J. Bacteriol. 2001, 183, 2834–2841. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Liu, Y.; Zheng, X.F.; Shang, K.; Cheng, M.L.; Wang, L.; Yang, N.; Yue, B.S. Characterization of olfactory receptor repertoires provides insights into the high-altitude adaptation of the yak based on the chromosome-level genome. Int. J. Biol. Macromol. 2022, 209, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Vats, P.; Singh, V.K.; Singh, S.N.; Singh, S.B. Glutathione Metabolism Under High-Altitude Stress and Effect of Antioxidant Supplementation. Aviat. Space Environ. Med. 2008, 79, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- Barata, H.; de Meis, L. Uncoupled ATP Hydrolysis and Thermogenic Activity of the Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 2002, 277, 16868–16872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, D.E.; Vouret-Craviari, V.; Pouyssegur, J. Angiogenesis and G-protein-coupled receptors: Signals that bridge the gap. Oncogene 2001, 20, 1556–1562. [Google Scholar] [CrossRef]
- Rawal, S.B.; Singh, M.V.; Tyagi, A.K.; Roy, J.; Dimri, G.P.; Selvamurthy, W. Effect of time exposure to high altitude on zinc and copper concentrations in human plasma. Aviat. Space Environ. Med. 1999, 70, 1161–1165. [Google Scholar]
- Voelkel, N.F.; Morris, K.G.; McMurtry, I.F.; Reeves, O.T. Calcium augments hypoxic vasoconstriction in lungs from high-altitude rats. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1980, 3, 450–455. [Google Scholar] [CrossRef]
- Aceituno-Madera, P.; Buendía-Eisman, A.; Olmo, F.J.; Jiménez-Moleón, J.J.; Serrano-Ortega, S. Melanoma, altitude, and UV-B radiation. Actas Dermo-Sifiliogr. 2011, 102, 199–205. [Google Scholar] [CrossRef]
- Hui, A.S.; Bauer, A.L.; Striet, J.B.; Schnell, P.O.; Czyzyk Krzeska, M.F. Calcium signaling stimulates translation of HIF-α during hypoxia. FASEB J. 2006, 20, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Sang, N.; Stiehl, D.P.; Bohensky, J.; Leshchinsky, I.; Srinivas, V.; Caro, J. MAPK Signaling Up-regulates the Activity of Hypoxia-inducible Factors by Its Effects on p300. J. Biol. Chem. 2003, 278, 14013–14019. [Google Scholar] [CrossRef] [Green Version]
- Yanagawa, Y.; Komatsu, S. Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots, independent of oxygen limitation. Plant Sci. 2012, 185–186, 250–258. [Google Scholar] [CrossRef]
- Zieseniss, A. Hypoxia and the modulation of the actin cytoskeleton –emerging interrelations. Hypoxia 2014, 2, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.M.; Zhang, X.J.; Liu, X.X.; Zhang, G.; Shang, W.Y.; Xue, J.; Chen, R.; Xing, Y.; Song, D.G.; Xu, R.H. Ginsenoside Rg1 promotes cerebral angiogenesis via the PI3K/Akt/mTOR signaling pathway in ischemic mice. Eur. J. Pharmacol. 2019, 856, 172418. [Google Scholar] [CrossRef] [PubMed]
- Kranenburg, O.; Gebbink, M.F.B.G.; Voest, E.E. Stimulation of angiogenesis by Ras proteins. Biochim. Biophys. Acta 2004, 1654, 23–37. [Google Scholar] [CrossRef] [PubMed]
- van Cruijsen, H.; Giaccone, G.; Hoekman, K. Epidermal growth factor receptor and angiogenesis: Opportunities for combined anticancer strategies. Int. J. Cancer. 2005, 117, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, S.J.; James, D.E.; Mann, M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef]
- Ke, Q.M.; Wu, J.; Tian, L.; Li, W.; Du, Y.M. Role of voltage-gated potassium channels in pathogenesis of chronic pulmonary heart disease. J. Huazhong Univ. Sci. Technol. [Med. Sci.] 2013, 33, 644–649. [Google Scholar] [CrossRef]
- Mishra, O.P.; Delivoria-Papadopoulos, M. Effect of hypoxia on protein tyrosine kinase activity in cortical membranes of newborn piglets—The role of nitric oxide. Neurosci. Lett. 2004, 372, 114–118. [Google Scholar] [CrossRef]
- Nakayama, K. Cellular Signal Transduction of the Hypoxia Response. J. Biochem. 2009, 146, 757–765. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol. Cancer. 2020, 19, 164. [Google Scholar] [CrossRef]
- Xia, J.; Liu, G.; Chen, Z.; Mao, C.; Zhou, D.; Wu, H.; Park, K.; Zhao, H.; Kim, S.; Cai, D.; et al. Hypoxia/ischemia promotes CXCL10 expression in cardiac microvascular endothelial cells by NFkB activation. Cytokine 2016, 81, 63–70. [Google Scholar] [CrossRef]
- Yang, S.; Lee, K.T.; Lee, J.Y.; Lee, J.K.; Lee, K.H.; Rhee, J.C. Inhibition of SCAMP1 suppresses cell migration and invasion in human pancreatic and gallbladder cancer cells. Tumor Biol. 2013, 34, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Jha, K.N. High Altitude and the Eye. Asia Pac. J. Ophthalmol. 2012, 1, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Takemura, Y.; Satoh, M.; Ozaki, K.; Kubota, S. Loss of FYCO1 leads to cataract formation. Sci. Rep. 2021, 11, 13771. [Google Scholar] [CrossRef] [PubMed]


| Abbreviation | Full Name | Function |
|---|---|---|
| ERBIN | Erbb2 interacting protein | Enhanced the expression of Hif-1α protein and promoted angiogenesis |
| CXCL10 | C-X-C motif chemokine | Hypoxia response related gene |
| SCAMP1 | Secretory carrier membrane protein 1 | Vascular endothelial growth factor related gene |
| FYCO1 | FYVE and coiled-coil domain-containing protein 1 | UV-radiation damage repair related gene |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, T.; Zhou, S.; Fang, J.; Wang, L.; Shi, L.; Dong, Y.; Zhang, H. Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals. Animals 2022, 12, 3572. https://doi.org/10.3390/ani12243572
Lyu T, Zhou S, Fang J, Wang L, Shi L, Dong Y, Zhang H. Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals. Animals. 2022; 12(24):3572. https://doi.org/10.3390/ani12243572
Chicago/Turabian StyleLyu, Tianshu, Shengyang Zhou, Jiaohui Fang, Lidong Wang, Lupeng Shi, Yuehuan Dong, and Honghai Zhang. 2022. "Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals" Animals 12, no. 24: 3572. https://doi.org/10.3390/ani12243572
APA StyleLyu, T., Zhou, S., Fang, J., Wang, L., Shi, L., Dong, Y., & Zhang, H. (2022). Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals. Animals, 12(24), 3572. https://doi.org/10.3390/ani12243572






