Genomic Characterization of hox Genes in Senegalese Sole (Solea senegalensis, Kaup 1858): Clues to Evolutionary Path in Pleuronectiformes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. PCR Screening of hox Genes in a BAC Genomic Library of Solea senegalensis
2.2. Extraction and Quantification of BAC Clones
2.3. BAC Sequencing and Bioinformatics Analysis
2.4. Analysis of Repetitive Elements
2.5. Comparative Genomics
2.6. BAC-FISH Mapping
2.7. Phylogenetic Analysis
3. Results
3.1. BAC Sequencing and Annotation
3.2. Analysis of Repetitive Elements
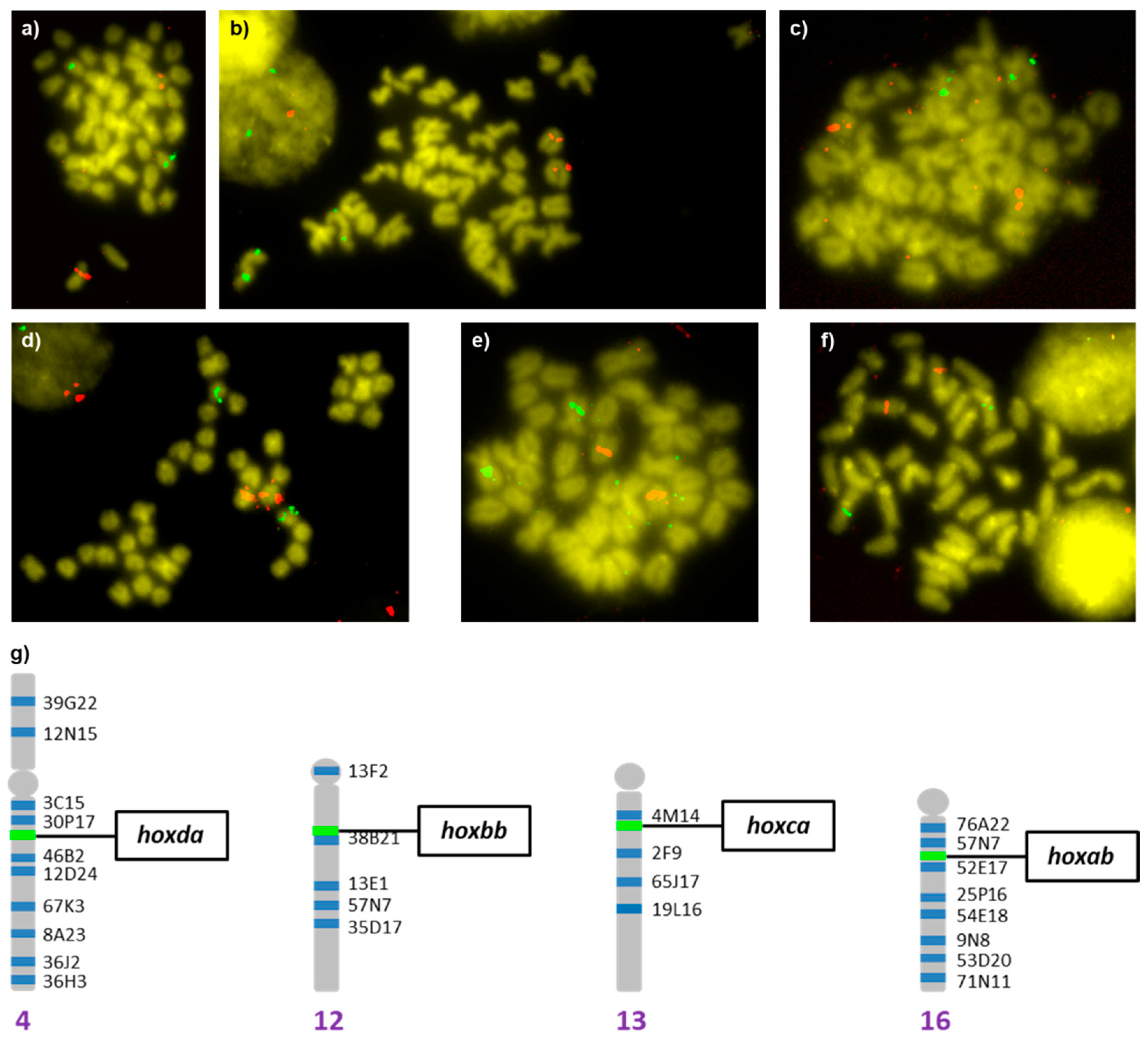
3.3. Cytogenetic Localization
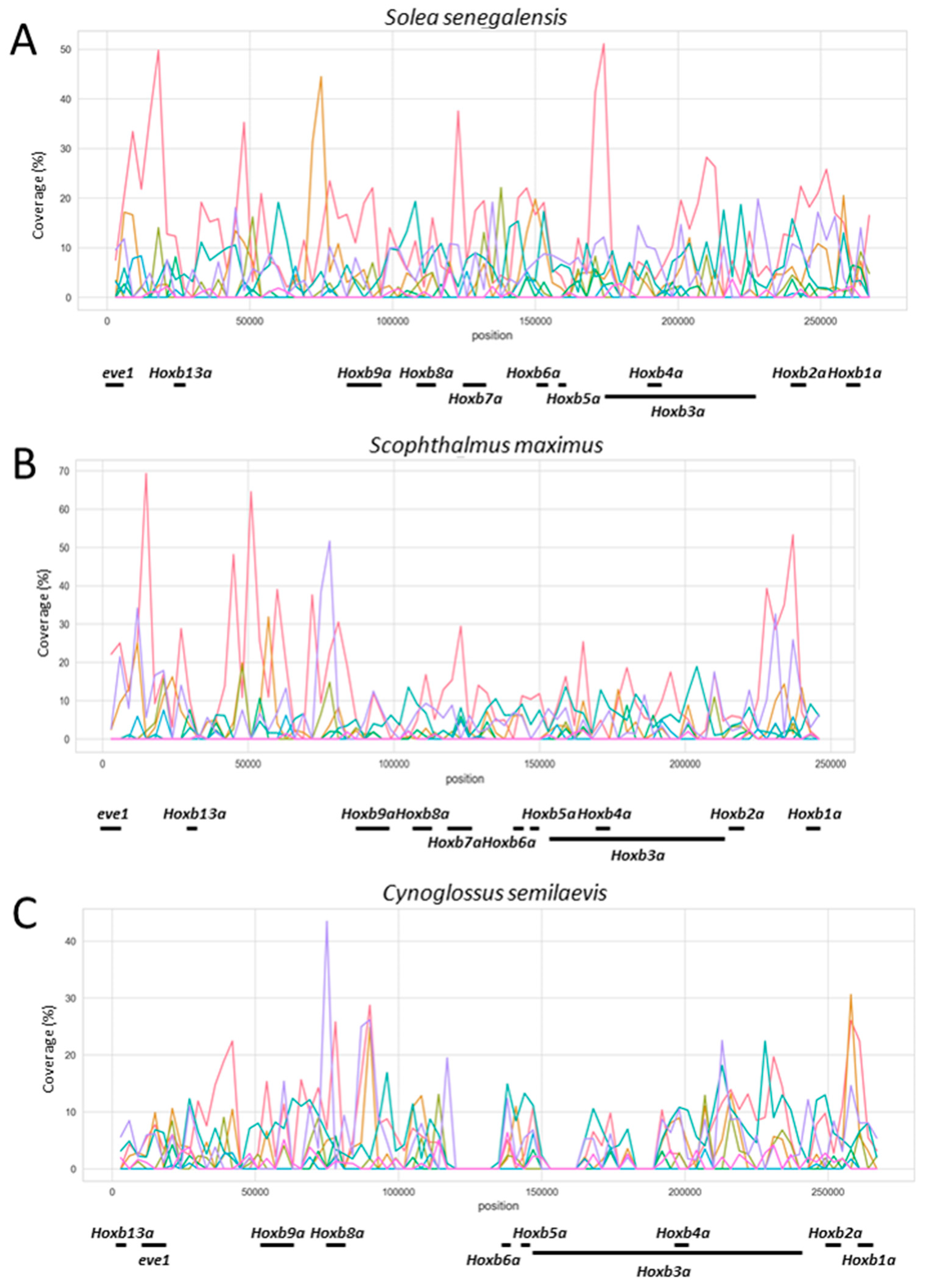
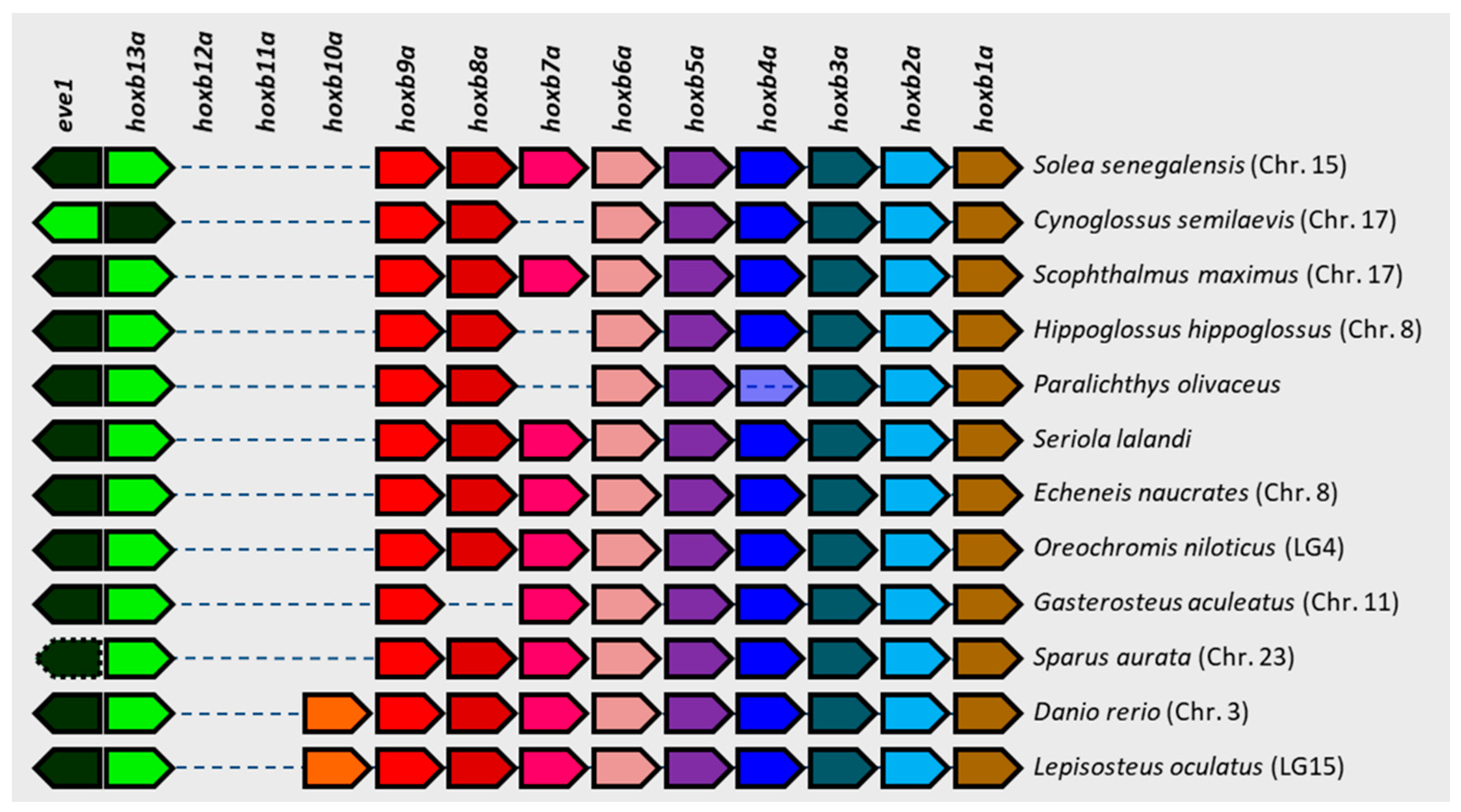
3.4. Comparative Genomics
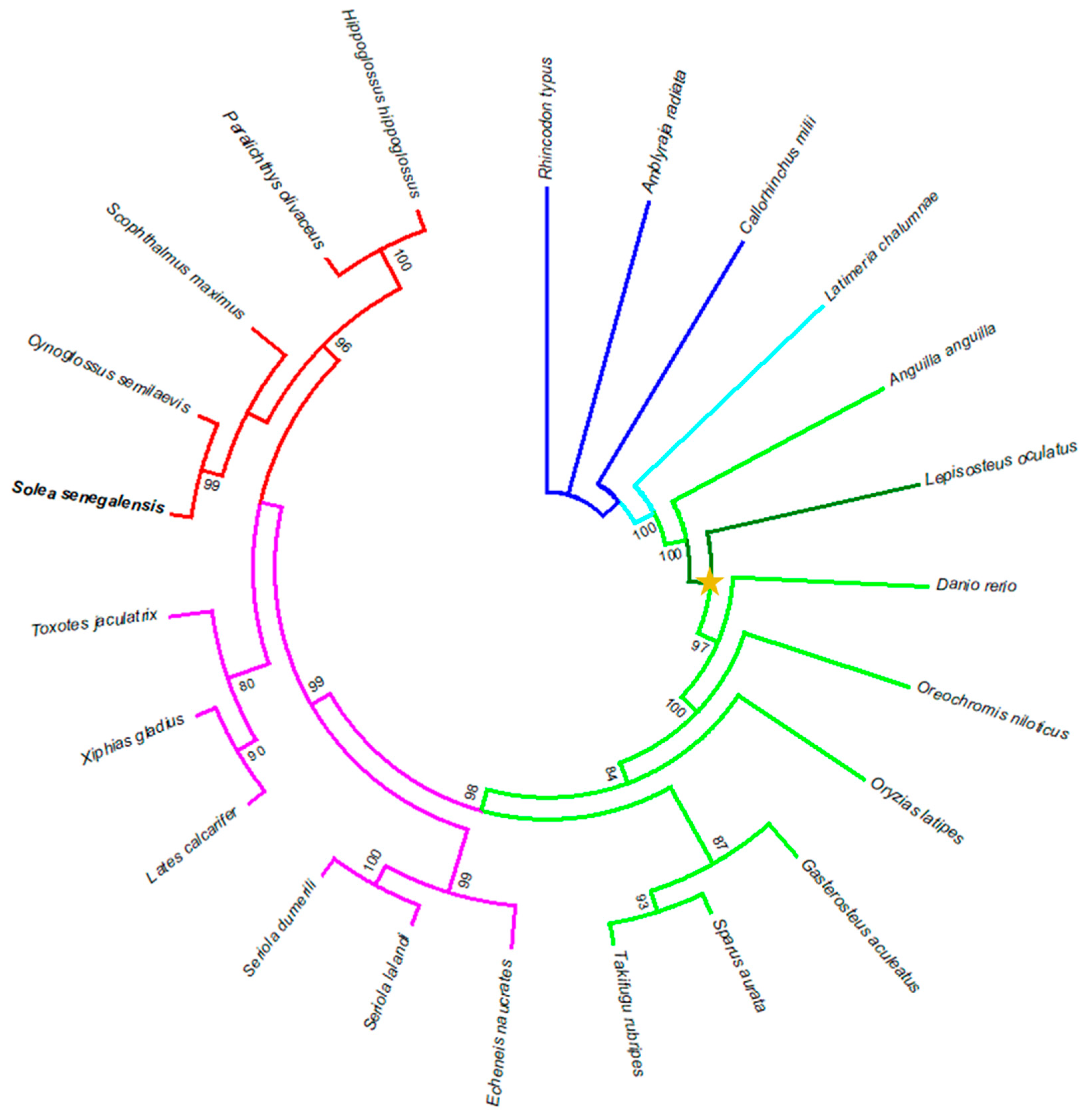
3.5. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Imsland, A.K.; Foss, A.; Dinis, M.T.; Delbare, D.; Schram, E.; Kamstra, A.; Rema, P.; White, P. A review of the culture potential of Solea solea and S. senegalensis. Rev. Fish Biol. Fish. 2004, 13, 379–407. [Google Scholar] [CrossRef]
- Morais, S.; Aragão, C.; Cabrita, E.; Conceição, L.E.; Constenla, M.; Costas, B.; Dias, J.; Duncan, N.; Engrola, S.; Estevez, A.; et al. New developments and biological insights into the farming of Solea senegalensis reinforcing its aquaculture potential. Rev. Aquac. 2014, 8, 227–263. [Google Scholar] [CrossRef]
- Cerdà, J.; Manchado, M. Advances in genomics for flatfish aquaculture. Genes Nutr. 2012, 8, 5–17. [Google Scholar] [CrossRef]
- Diopere, E.; Maes, G.E.; Komen, H.; Volckaert, F.A.M.; Groenen, M.A.M. A Genetic Linkage Map of Sole (Solea solea): A Tool for Evolutionary and Comparative Analyses of Exploited (Flat)Fishes. PLoS ONE 2014, 9, e115040. [Google Scholar] [CrossRef]
- Guerrero-Cózar, I.; Perez-Garcia, C.; Benzekri, H.; Sánchez, J.J.; Seoane, P.; Cruz, F.; Gut, M.; Zamorano, M.J.; Claros, M.G.; Manchado, M. Development of whole-genome multiplex assays and construction of an integrated genetic map using SSR markers in Senegalese sole. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Vega, L.; Díaz, E.; Cross, I.; Rebordinos, L. Caracterizaciones citogenética e isoenzimática del lenguado S. senegalensis Kaup, 1858. Bol. IEO 2002, 18, 245–250. [Google Scholar]
- Cross, I.; Merlo, A.; Manchado, M.; Infante, C.; Cañavate, J.P.; Rebordinos, L.; Torres, M.A.M. Cytogenetic characterization of the sole Solea senegalensis (Teleostei: Pleuronectiformes: Soleidae): Ag-NOR, (GATA) n, (TTAGGG) n and ribosomal genes by one-color and two-color FISH. Genetica 2006, 128, 253–259. [Google Scholar] [CrossRef]
- Rodríguez, M.E.; Molina, B.; Merlo, M.A.; Arias-Pérez, A.; Portela-Bens, S.; García-Angulo, A.; Cross, I.; Liehr, T.; Rebordinos, L. Evolution of the Proto Sex-Chromosome in Solea senegalensis. Int. J. Mol. Sci. 2019, 20, 5111. [Google Scholar] [CrossRef] [PubMed]
- Cross, I.; García, E.; Rodríguez, M.E.; Arias-Pérez, A.; Portela-Bens, S.; Merlo, M.A.; Rebordinos, L. The genomic structure of the highly-conserved dmrt1 gene in Solea senegalensis (Kaup, 1868) shows an unexpected intragenic duplication. PLoS ONE 2020, 15, e0241518. [Google Scholar] [CrossRef]
- García-Angulo, A.; Merlo, M.A.; Portela-Bens, S.; Rodríguez, M.E.; García, E.; Al-Rikabi, A.; Liehr, T.; Rebordinos, L. Evidence for a Robertsonian fusion in Solea senegalensis (Kaup, 1858) revealed by zoo-FISH and comparative genome analysis. BMC Genom. 2018, 19, 818. [Google Scholar] [CrossRef]
- García-Angulo, A.; Merlo, M.A.; Rodríguez, M.E.; Portela-Bens, S.; Liehr, T.; Rebordinos, L. Genome and Phylogenetic Analysis of Genes Involved in the Immune System of Solea senegalensis—Potential Applications in Aquaculture. Front. Genet. 2019, 10, 529. [Google Scholar] [CrossRef]
- García-Angulo, A.; Merlo, M.A.; Iziga, R.; Rodríguez, M.E.; Portela-Bens, S.; Al-Rikabi, A.; Liehr, T.; Rebordinos, L. Gene clusters related to metamorphosis in Solea senegalensis are highly conserved. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100706. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Cross, I.; Arias-Pérez, A.; Portela-Bens, S.; Merlo, M.; Liehr, T.; Rebordinos, L. Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis. Int. J. Mol. Sci. 2021, 22, 1614. [Google Scholar] [CrossRef] [PubMed]
- Merlo, M.A.; Iziga, R.; Portela-Bens, S.; Cross, I.; Kosyakova, N.; Liehr, T.; Manchado, M.; Rebordinos, L. Analysis of the histone cluster in Senegalese sole (Solea senegalensis): Evidence for a divergent evolution of two canonical histone clusters. Genome 2017, 60, 441–453. [Google Scholar] [CrossRef] [PubMed]
- García, E.; Cross, I.; Portela-Bens, S.; Rodríguez, M.E.; García-Angulo, A.; Molina, B.; Cuadrado, A.; Liehr, T.; Rebordinos, L. Integrative genetic map of repetitive DNA in the sole Solea senegalensis genome shows a Rex transposon located in a proto-sex chromosome. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Merlo, M.; Portela-Bens, S.; Rodríguez, M.; García-Angulo, A.; Cross, I.; Arias-Pérez, A.; García, E.; Rebordinos, L. A Comprehensive Integrated Genetic Map of the Complete Karyotype of Solea senegalensis (Kaup 1858). Genes 2021, 12, 49. [Google Scholar] [CrossRef]
- Ramírez, D.; Rodríguez, M.E.; Cross, I.; Arias-Pérez, A.; Merlo, M.A.; Anaya, M.; Portela-Bens, S.; Martínez, P.; Robles, F.; Ruiz-Rejón, C.; et al. Integration of Maps Enables a Cytogenomics Analysis of the Complete Karyotype in Solea senegalensis. Int. J. Mol. Sci. 2022, 23, 5353. [Google Scholar] [CrossRef]
- Gibson, R.N.; Nash, R.D.M.; Geffen, A.J.; Van Der Veer, H.W. (Eds.) Flatfishes: Biology and Exploitation, 2nd ed.; John Wiley & Sons, Incorporated: Oxford, UK, 2015; pp. 74–166. [Google Scholar] [CrossRef]
- Padrós, F.; Villalta, M.; Gisbert, E.; Estévez, A. Morphological and histological study of larval development of the Senegal sole Solea senegalensis: An integrative study. J. Fish Biol. 2011, 79, 3–32. [Google Scholar] [CrossRef]
- Fernández, I.; Granadeiro, L.; Darias, M.J.; Gavaia, P.J.; Andree, K.B.; Gisbert, E. Solea senegalensis skeletal ossification and gene expression patterns during metamorphosis: New clues on the onset of skeletal deformities during larval to juvenile transition. Aquaculture 2018, 496, 153–165. [Google Scholar] [CrossRef]
- Muñoz-Cueto, J.A.; Mañanos Sanchez, E.L.; Sanchez Vasquez, F.J. The Biology of Sole; CRC Press: Boca Raton, FL, USA, 2019; pp. 216–252. [Google Scholar]
- Mallo, M. The vertebrate tail: A gene playground for evolution. Cell Mol. Life Sci. 2020, 77, 1021–1030. [Google Scholar] [CrossRef]
- Mallo, M. Of Necks, Trunks and Tails: Axial Skeletal Diversity among Vertebrates. Diversity 2021, 13, 289. [Google Scholar] [CrossRef]
- Tanaka, M. Fins into limbs: Autopod acquisition and anterior elements reduction by modifying gene networks involving 5′Hox, Gli3, and Shh. Dev. Biol. 2016, 413, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, L.B.; Bikoff, J.B.; Gabitto, M.I.; Brenner-Morton, S.; Baek, M.; Yang, J.H.; Tabak, E.G.; Dasen, J.S.; Kintner, C.R.; Jessell, T.M. Origin and Segmental Diversity of Spinal Inhibitory Interneurons. Neuron 2018, 97, 341–355.e3. [Google Scholar] [CrossRef]
- Archambeault, S.; Taylor, J.A.; Crow, K.D. HoxA and HoxD expression in a variety of vertebrate body plan features reveals an ancient origin for the distal Hox program. Evodevo 2014, 5, 44. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dietrich, K.; Fiedler, I.A.; Kurzyukova, A.; López-Delgado, A.C.; McGowan, L.M.; Geurtzen, K.; Hammond, C.L.; Busse, B.; Knopf, F. Skeletal Biology and Disease Modeling in Zebrafish. J. Bone Miner. Res. 2021, 36, 436–458. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.B.; Henke, K.; Harris, M.P. Latent developmental potential to form limb-like skeletal structures in zebrafish. Cell 2021, 184, 899–911.e13. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.; Ho, R.K. Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: Implications for the evolution of vertebrate paired appendages. Dev. Biol. 2008, 322, 220–233. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Yao, X.; Gao, F.; Bao, B. Dorsal fin development in flounder, Paralichthys olivaceus: Bud formation and its cellular origin. Gene Expr. Patterns 2017, 25–26, 22–28. [Google Scholar] [CrossRef]
- Davis, M.C.; Dahn, R.D.; Shubin, N.H. An autopodial-like pattern of Hox expression in the fins of a basal actinopterygian fish. Nature 2007, 447, 473–476. [Google Scholar] [CrossRef]
- Brown, J.M.; Travers, S.K.; Dodgen, C.D.; Duffus AL, J.; Davis, A. Gene expression pattern analysis of anterior hox genes during zebrafish (Danio rerio) embryonic development reveals divergent expression patterns from other teleosts. Ga. J. Sci. 2020, 78, 1–12. [Google Scholar]
- Suzuki, T.; Oohara, I.; Kurokawa, T. Retinoic acid given at late embryonic stage depresses sonic hedgehog and Hoxd-4 expression in the pharyngeal area and induces skeletal malformation in flounder (Paralichthys olivaceus) embryos. Dev. Growth Differ. 1999, 41, 143–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davis, A.; Stellwag, E.J. Spatio-temporal patterns of Hox paralog group 3–6 gene expression during Japanese medaka (Oryzias latipes) embryonic development. Gene Expr. Patterns 2010, 10, 244–250. [Google Scholar] [CrossRef]
- Lyon, R.S.; Davis, A.; Scemama, J.-L. Spatio-temporal expression patterns of anterior Hox genes during Nile tilapia (Oreochromis niloticus) embryonic development. Gene Expr. Patterns 2013, 13, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.J.; Holland, P.W.H. Diversification of Hox Gene Clusters in Osteoglossomorph Fish in Comparison to Other Teleosts and the Spotted Gar Outgroup. J. Exp. Zool. Part B Mol. Dev. Evol. 2017, 328, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fernandez, J. Hox, ParaHox, ProtoHox: Facts and guesses. Heredity 2005, 94, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Q.; Huang, X.; Yang, C.; Chen, L.; Han, M.; Shu, Y.; Wang, M.; Li, W.; Hu, F.; et al. The rapid variation of Hox clusters reveals a clear evolutionary path in a crucian carp-like homodiploid fish lineage. Reprod. Breed. 2021, 1, 149–156. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, Y.; Zou, L.; Luo, Y.; Tan, H.; Yao, J.; Zhang, M.; Liu, S. Hox genes reveal variations in the genomic DNA of allotetraploid hybrids derived from Carassius auratus red var. (female) × Cyprinus carpio L. (male). BMC Genet. 2020, 21, 24. [Google Scholar] [CrossRef]
- Yamada, K.; Maeno, A.; Araki, S.; Kikuchi, M.; Suzuki, M.; Ishizaka, M.; Satoh, K.; Akama, K.; Kawabe, Y.; Suzuki, K.; et al. An atlas of seven zebrafish hox cluster mutants provides insights into sub/neofunctionalization of vertebrate Hox clusters. Development 2021, 148, dev198325. [Google Scholar] [CrossRef]
- Kavouras, M.; Malandrakis, E.E.; Golomazou, E.; Konstantinidis, I.; Blom, E.; Palstra, A.P.; Anastassiadis, K.; Panagiotaki, P.; Exadactylos, A. Hox gene expression profiles during embryonic development of common sole. Anim. Biol. 2019, 69, 183–198. [Google Scholar] [CrossRef]
- Kavouras, M.; Malandrakis, E.E.; Danis, T.; Blom, E.; Anastassiadis, K.; Panagiotaki, P.; Exadactylos, A. Hox genes polymorphism depicts developmental disruption of common sole eggs. Open Life Sci. 2019, 14, 549–563. [Google Scholar] [CrossRef]
- Symonová, R.; Havelka, M.; Amemiya, C.T.; Howell, W.M.; Kořínková, T.; Flajšhans, M.; Gela, D.; Ráb, P. Molecular cytogenetic differentiation of paralogs of Hox paralogs in duplicated and re-diploidized genome of the North American paddlefish (Polyodon spathula). BMC Genet. 2017, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, S.; Abe, I.; Kudoh, Y.; Kishi, N.; Wang, Y.; Kubota, R.; Kudoh, J.; Kawasaki, K.; Minoshima, S.; Shimizu, N. Human BAC library: Construction and rapid screening. Gene 1997, 191, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Crow, K.D.; Stadler, P.F.; Lynch, V.J.; Amemiya, C.; Wagner, G.P. The “Fish-Specific” Hox Cluster Duplication Is Coincident with the Origin of Teleosts. Mol. Biol. Evol. 2006, 23, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. United States N. p. 2014. Available online: sourceforge.net/projects/bbmap/ (accessed on 1 May 2020).
- Ewing, B.; Green, P. Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef]
- Coelho, L.P.; Alves, R.; Monteiro, P.; Huerta-Cepas, J.; Freitas, A.T.; Bork, P. NG-meta-profiler: Fast processing of metagenomes using NGLess, a domain-specific language. Microbiome 2019, 7, 84. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University, State College, PA, USA, 2007. [Google Scholar]
- Portela-Bens, S.; Merlo, M.A.; Rodríguez, M.E.; Cross, I.; Manchado, M.; Kosyakova, N.; Liehr, T.; Rebordinos, L. Integrated gene mapping and synteny studies give insights into the evolution of a sex proto-chromosome in Solea senegalensis. Chromosoma 2017, 126, 261–277. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef]
- Xia, X. DAMBE6: New Tools for Microbial Genomics, Phylogenetics, and Molecular Evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Guerrero-Cózar, I.; Gomez-Garrido, J.; Berbel, C.; Martinez-Blanch, J.F.; Alioto, T.; Claros, M.G.; Gagnaire, P.-A.; Manchado, M. Chromosome anchoring in Senegalese sole (Solea senegalensis) reveals sex-associated markers and genome rearrangements in flatfish. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Covelo-Soto, L.; Saura, M.; Morán, P. Does DNA methylation regulate metamorphosis? The case of the sea lamprey (Petromyzon marinus) as an example. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 185, 42–46. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic Timing of the Fish-Specific Genome Duplication Correlates with the Diversification of Teleost Fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L.; et al. Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Kuraku, S. Hox Gene Clusters of Early Vertebrates: Do They Serve as Reliable Markers for Genome Evolution? Genom. Proteom. Bioinform. 2011, 9, 97–103. [Google Scholar] [CrossRef]
- Cheng, P.; Huang, Y.; Du, H.; Li, C.; Lv, Y.; Ruan, R.; Ye, H.; Bian, C.; You, X.; Xu, J.; et al. Draft Genome and Complete Hox-Cluster Characterization of the Sterlet (Acipenser ruthenus). Front. Genet. 2019, 10, 776. [Google Scholar] [CrossRef]
- Henkel, C.V.; Burgerhout, E.; de Wijze, D.L.; Dirks, R.P.; Minegishi, Y.; Jansen, H.J.; Spaink, H.P.; Dufour, S.; Weltzien, F.-A.; Tsukamoto, K.; et al. Primitive Duplicate Hox Clusters in the European Eel’s Genome. PLoS ONE 2012, 7, e32231. [Google Scholar] [CrossRef] [PubMed]
- Hurley, I.A.; Mueller, R.L.; Dunn, K.; Schmidt, E.J.; Friedman, M.; Ho, R.K.; Prince, V.; Yang, Z.; Thomas, M.; Coates, M.I. A new time-scale for ray-finned fish evolution. Proc. R. Soc. B Biol. Sci. 2007, 274, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.J.; Holland, P.W. Enigmatic Orthology Relationships between Hox Clusters of the African Butterfly Fish and Other Teleosts Following Ancient Whole-Genome Duplication. Mol. Biol. Evol. 2014, 31, 2592–2611. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-M.; Lee, B.-Y.; Lee, J.-H.; Rhee, J.-S. Conservation of Hox gene clusters in the self-fertilizing fish Kryptolebias marmoratus (Cyprinodontiformes; Rivulidae). J. Fish Biol. 2016, 88, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Hwang, D.-S.; Jeong, C.-B.; Au, D.W.T.; Lee, J.-S. Identification and conservation of gene loss events of Hox gene clusters in the marine medaka (Oryzias melastigma). J. Exp. Zool. Part B Mol. Dev. Evol. 2016, 326, 387–393. [Google Scholar] [CrossRef]
- Hoegg, S.; Boore, J.L.; Kuehl, J.V.; Meyer, A. Comparative phylogenomic analyses of teleost fish Hox gene clusters: Lessons from the cichlid fish Astatotilapia burtoni. BMC Genom. 2007, 8, 317. [Google Scholar] [CrossRef]
- Fried, C.; Prohaska, S.J.; Stadler, P.F. Exclusion of repetitive DNA elements from gnathostome Hox clusters. J. Exp. Zool. B Mol. Dev. Evol. 2004, 302, 165–173. [Google Scholar] [CrossRef]
- Feiner, N.; Wood, N.J. Lizards possess the most complete tetrapod Hox gene repertoire despite pervasive structural changes in Hox clusters. Evol. Dev. 2019, 21, 218–228. [Google Scholar] [CrossRef]
- Di-Poï, N.; Montoya-Burgos, J.I.; Duboule, D. Atypical relaxation of structural constraints in Hox gene clusters of the green anole lizard. Genome Res. 2009, 19, 602–610. [Google Scholar] [CrossRef]
- Simons, C.; Pheasant, M.; Makunin, I.V.; Mattick, J.S. Transposon-free regions in mammalian genomes. Genome Res. 2006, 16, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Simons, C.; Makunin, I.V.; Pheasant, M.; Mattick, J.S. Maintenance of transposon-free regions throughout vertebrate evolution. BMC Genom. 2007, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R.; Suh, A. Nucleotide composition of transposable elements likely contributes to AT/GC compositional homogeneity of teleost fish genomes. Mob. DNA 2019, 10, 49. [Google Scholar] [CrossRef]
- Feiner, N. Accumulation of transposable elements in Hox gene clusters during adaptive radiation of Anolis lizards. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161555. [Google Scholar] [CrossRef] [PubMed]
- Rochtus, A.; Izzi, B.; Vangeel, E.; Louwette, S.; Wittevrongel, C.; Lambrechts, D.; Moreau, Y.; Winand, R.; Verpoorten, C.; Jansen, K.; et al. DNA methylation analysis of Homeobox genes implicates HOXB7 hypomethylation as risk factor for neural tube defects. Epigenetics 2015, 10, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Ernst, P.; Wang, Y.; Dekens, M.P.S.; Kingsley, P.D.; Palis, J.; Korsmeyer, S.J.; Daley, G.Q.; Zon, L.I. Cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 2003, 425, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Corredor-Adamez, M.; Welten, M.C.M.; Spaink, H.P.; Jeffery, J.E.; Schoon, R.T.; de Bakker, M.A.G.; Bagowski, C.P.; Meijer, A.H.; Verbeek, F.J.; Richardson, M.K. Genomic annotation and transcriptome analysis of the zebrafish (Danio rerio) hox complex with description of a novel member, hoxb13a. Evol. Dev. 2005, 7, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C. The contribution of transposable elements at the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef]
- Zhao, G.; Zou, C.; Li, K.; Wang, K.; Li, T.; Gao, L.; Zhang, X.; Wang, H.; Yang, Z.; Liu, X.; et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 2017, 3, 946–955. [Google Scholar] [CrossRef]


| Loci | Forward Primer 5′–3′ | Reverse Primer 5′–3′ | Ta (°C) | Reference |
|---|---|---|---|---|
| hoxab | CGGGCGAGAGAGTGGTTTATCAA | CGGAGTATCCGTGGATGAAGGAGA | 64 | Present work |
| hoxbb | TAYCCRAATGGSYCYGACTA | TYCKCATCCARGGRAAWATYTG | 66 | [45] |
| hoxca | GACCACGGGTCCCATAAGTAAT | CTCATGTCAGTGGATGAGCAGT | 54 | Present work |
| hoxda | CCAAACGGTCCAGAACAGCTTA | CAGTCTGCCCTTGGTGTTGG | 64 | Present work |
| Cluster | Best-Fit Model | Decoration | AIC | lnL |
|---|---|---|---|---|
| hoxaa | TN93 | +R | 40,758.16648 | −20,299.94113 |
| hoxab | TN93 | +R | 36,039.57872 | −17,927.10111 |
| hoxba | GTR | +R | 46,753.97854 | −23,296.59684 |
| hoxbb | TN93 | +R | 34,577.30982 | −17,194.72750 |
| hoxca | TN93 | +R | 23,974.40940 | −11,912.02291 |
| hoxda | GTR | +R | 36,446.84214 | −18,078.12721 |
| hoxdb | GTR | +R | 31,505.40632 | −15,666.28770 |
| Cluster | BAC | Total Length (bp) | N50 (bp) | L50 | Annotated Genes | Accession Number |
|---|---|---|---|---|---|---|
| hoxab | 6D19 | 256,967 | 30,145 | 4 | snx10b, skap2, hoxa2b, hoxa9b, hoxa10b, hoxa11b, hoxa13b, hibadhb, tax1bp1b, jazf1b, creb5b, chn2, cpvl | OK504498 |
| hoxbb | 39L15 | 192,932 | 50,613 | 2 | skap1, hoxb1b, hoxb3b, hoxb5b, hoxb6b, ttll6, calcoco2, snf8, msl1b, chmp2a, mrm1, fbrs, srcap, stx4, arhgap44 | OK474316 |
| hoxca | 51K20 | 200,678 | 20,233 | 3 | hoxc3a, hoxc4a, hoxc5a, hoxc6a, hoxc8a, hoxc9a, hoxc10a, hoxc11a, hoxc12a, hoxc13a, calcoco1a, rarga | OK474317 |
| hoxda | 29F20 | 159,784 | 45,033 | 2 | hoxd4a, hoxd3a | OK504499 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendizábal-Castillero, M.; Merlo, M.A.; Cross, I.; Rodríguez, M.E.; Rebordinos, L. Genomic Characterization of hox Genes in Senegalese Sole (Solea senegalensis, Kaup 1858): Clues to Evolutionary Path in Pleuronectiformes. Animals 2022, 12, 3586. https://doi.org/10.3390/ani12243586
Mendizábal-Castillero M, Merlo MA, Cross I, Rodríguez ME, Rebordinos L. Genomic Characterization of hox Genes in Senegalese Sole (Solea senegalensis, Kaup 1858): Clues to Evolutionary Path in Pleuronectiformes. Animals. 2022; 12(24):3586. https://doi.org/10.3390/ani12243586
Chicago/Turabian StyleMendizábal-Castillero, Marco, Manuel Alejandro Merlo, Ismael Cross, María Esther Rodríguez, and Laureana Rebordinos. 2022. "Genomic Characterization of hox Genes in Senegalese Sole (Solea senegalensis, Kaup 1858): Clues to Evolutionary Path in Pleuronectiformes" Animals 12, no. 24: 3586. https://doi.org/10.3390/ani12243586
APA StyleMendizábal-Castillero, M., Merlo, M. A., Cross, I., Rodríguez, M. E., & Rebordinos, L. (2022). Genomic Characterization of hox Genes in Senegalese Sole (Solea senegalensis, Kaup 1858): Clues to Evolutionary Path in Pleuronectiformes. Animals, 12(24), 3586. https://doi.org/10.3390/ani12243586







