Characterization of Virulence Factors in Enterotoxin-Producing Staphylococcus aureus from Bulk Tank Milk
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. Detection of Virulence Factors
2.3. Molecular Typing
2.4. Statistical Analysis
3. Results
3.1. Prevalence of S. aureus with Enterotoxingenes
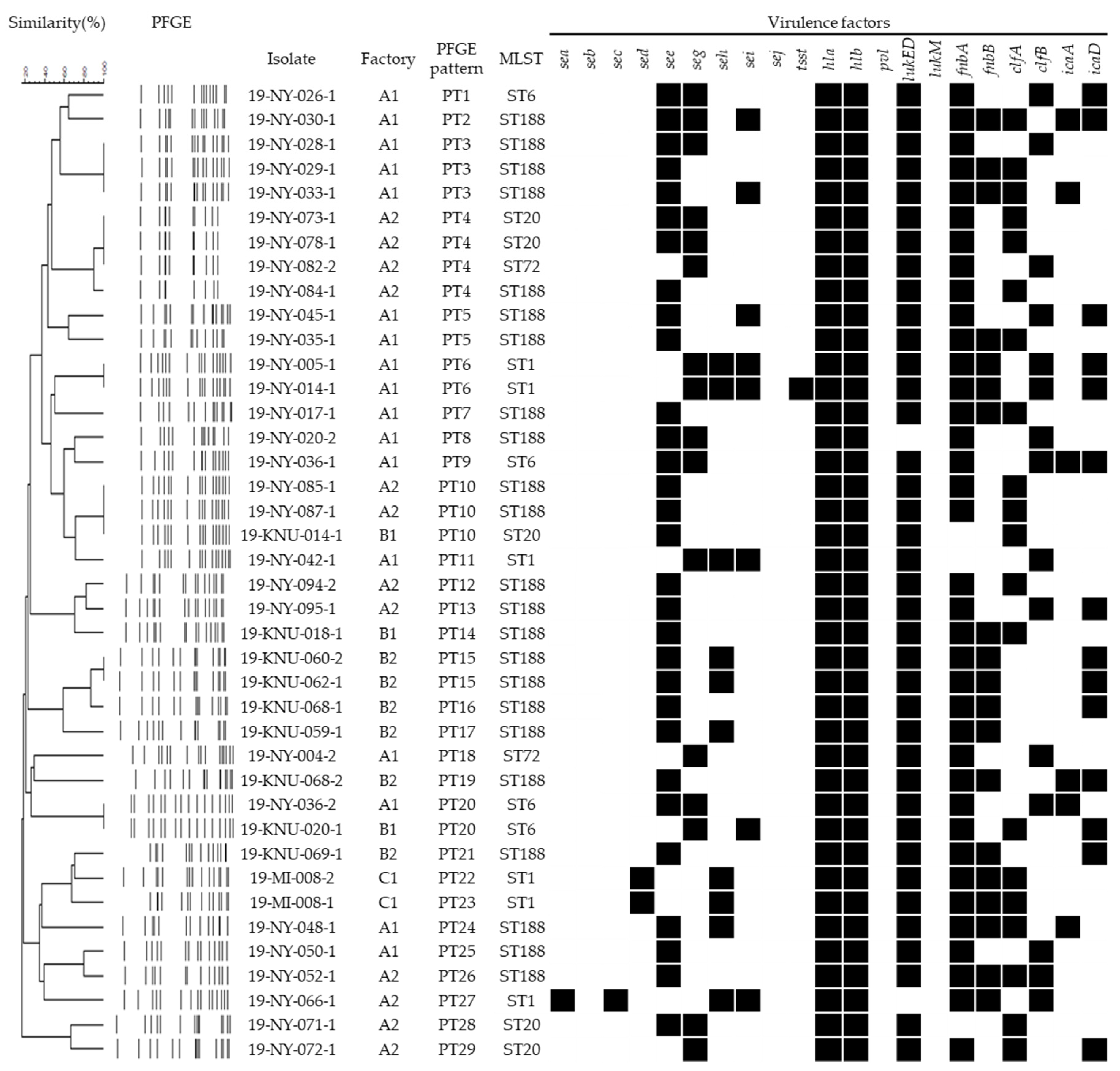
3.2. Distribution of Virulence Genes
3.3. Genotypic Characteristics of S. aureus Carrying the Enterotoxin Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monistero, V.; Graber, H.; Pollera, C.; Cremonesi, P.; Castiglioni, B.; Bottini, E.; Ceballos-Marquez, A.; Lasso-Rojas, L.; Kroemker, V.; Wente, N.; et al. Staphylococcus aureus Isolates from Bovine Mastitis in Eight Countries: Genotypes, Detection of Genes Encoding Different Toxins and Other Virulence Genes. Toxins 2018, 10, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereyra, E.A.L.; Picech, F.; Renna, M.S.; Baravalle, C.; Andreotti, C.S.; Russi, R.; Calvinho, L.F.; Diez, C.; Dallard, B.E. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet. Microbiol. 2016, 183, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Putz, E.J.; Palmer, M.V.; Ma, H.; Casas, E.; Reinhardt, T.A.; Lippolis, J.D. Case report: Characterization of a persistent, treatment-resistant, novel Staphylococcus aureus infection causing chronic mastitis in a Holstein dairy cow. BMC Vet. Res. 2020, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Krakauer, T. Staphylococcal superantigens: Pyrogenic toxins induce toxic shock. Toxins 2019, 11, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillaspy, A.F.; Iandolo, J.J. Staphylococcus: Introduction. In Encyclopedia of Food Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 3, pp. 482–486. [Google Scholar] [CrossRef]
- Pérez, V.K.C.; da Costa, G.M.; Guimarães, A.S.; Heinemann, M.B.; Lage, A.P.; Dorneles, E.M.S. Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. J. Glob. Antimicrob. Resist. 2020, 22, 792–802. [Google Scholar] [CrossRef]
- Homsombat, T.; Boonyayatra, S.; Awaiwanont, N.; Pichpol, D. Effect of temperature on the expression of classical enterotoxin genes among staphylococci associated with bovine mastitis. Pathogens 2021, 10, 975. [Google Scholar] [CrossRef]
- Lina, G.; Piémont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef]
- Lo, W.T.; Wang, C.C. Panton-valentine leukocidin in the pathogenesis of community-associated methicillin-resistant staphylococcus aureus infection. Pediatr. Neonatol. 2011, 52, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Ministry of Food and Drug Safety (MFDS). Processing Standards and Ingredient Specifications for Livestock Products; Ministry of Food and Drug Safety: Cheongju, Korea, 2018. [Google Scholar]
- Igbinosa, E.O.; Beshiru, A.; Akporehe, L.U.; Oviasogie, F.E.; Igbinosa, O.O. Prevalence of methicillin-resistant Staphylococcus aureus and other Staphylococcus species in raw meat samples intended for human consumption in Benin City, Nigeria: Implications for public health. Int. J. Environ. Res. Public Health 2016, 13, 949. [Google Scholar] [CrossRef]
- Faridi, A.; Kareshk, A.T.; Fatahi-Bafghi, M.; Ziasistani, M.; Ghahraman, M.R.K.; Seyyed-Yousefi, S.Z.; Shakeri, N.; Kalantar-Neyestanaki, D. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in clinical samples of patients with external ocular infection. Iran. J. Microbiol. 2018, 10, 215–219. [Google Scholar]
- Mashouf, R.Y.; Hosseini, S.M.; Mousavi, S.M.; Arabestani, M.R. Prevalence of enterotoxin genes and antibacterial susceptibility pattern of Staphylococcus aureus strains isolated from animal originated foods in West of Iran. Oman Med. J. 2015, 30, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Koosha, R.Z.; Hosseini, H.M.; Aghdam, E.M.; Fooladi, A.A.I.; Tajandareh, S.G. Distribution of tsst-1 and mecA genes in Staphylococcus aureus isolated from clinical specimens. Jundishapur J. Microbiol. 2016, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, A.; Chothe, S.; Byukusenge, M.; Mathews, T.; Pierre, T.; Kariyawasam, S.; Luley, E.; Kuchipudi, S.; Jayarao, B. Prevalence and distribution of multilocus sequence types of Staphylococcus aureus isolated from bulk tank milk and cows with mastitis in Pennsylvania. PLoS ONE 2021, 16, e0248528. [Google Scholar] [CrossRef]
- Xu, J.; Tan, X.; Zhang, X.; Xia, X.; Sun, H. The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb. Pathog. 2015, 88, 29–38. [Google Scholar] [CrossRef]
- Felipe, V.; Morgante, C.A.; Somale, P.S.; Varroni, F.; Zingaretti, M.L.; Bachetti, R.A.; Correa, S.G.; Porporatto, C. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb. Pathog. 2017, 104, 278–286. [Google Scholar] [CrossRef]
- Vasudevan, P.; Nair, M.K.M.; Annamalai, T.; Venkitanarayanan, K.S. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 2003, 92, 179–185. [Google Scholar] [CrossRef]
- Saunders, N.A.; Holmes, A. Multilocus sequence typing (MLST) of staphylococcus aureus. Methods Mol. Biol. 2014, 1085, 113–130. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention(CDC). Centers for Disease Control and Prevention; Centers for Disease Control and Prevention(CDC): Atlanta, GA, USA, 2020; ISBN 1115430610. [Google Scholar]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-Field Gel Electrophoresis Typing of Oxacillin-Resistant Staphylococcus aureus Isolates from the United States: Establishing a National Database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef] [Green Version]
- Bien, J.; Sokolova, O.; Bozko, P. Characterization of Virulence Factors of Staphylococcus aureus: Novel Function of Known Virulence Factors That Are Implicated in Activation of Airway Epithelial Proinflammatory Response. J. Pathog. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korea Agro-Fisheries & Food Trade Corporation Food Information Statistics System (ATFIS). Available online: https://www.atfis.or.kr/ (accessed on 1 December 2021).
- Peles, F.; Wagner, M.; Varga, L.; Hein, I.; Rieck, P.; Gutser, K.; Keresztúri, P.; Kardos, G.; Turcsányi, I.; Béri, B.; et al. Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. Int. J. Food Microbiol. 2007, 118, 186–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schelin, J.; Wallin-Carlquist, N.; Cohn, M.T.; Lindqvist, R.; Barker, G.C.; Rådström, P. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2011, 2, 580–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Zhang, L.; Zhou, X.; He, Y.; Yong, C.; Shen, M.; Szenci, O.; Han, B. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J. Dairy Sci. 2016, 99, 9560–9569. [Google Scholar] [CrossRef]
- Zaatout, N.; Ayachi, A.; Kecha, M.; Kadlec, K. Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J. Appl. Microbiol. 2019, 127, 1305–1314. [Google Scholar] [CrossRef]
- Liu, K.; Tao, L.; Li, J.; Fang, L.; Cui, L.; Li, J.; Meng, X.; Zhu, G.; Bi, C.; Wang, H. Characterization of Staphylococcus aureus Isolates From Cases of Clinical Bovine Mastitis on Large-Scale Chinese Dairy Farms. Front. Vet. Sci. 2020, 7, 1–9. [Google Scholar] [CrossRef]
- Löffler, B.; Hussain, M.; Grundmeier, M.; Brück, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peters, G. Staphylococcus aureus Panton-Valentine Leukocidin Is a Very Potent Cytotoxic Factor for Human Neutrophils. PLoS Pathog. 2010, 6, e1000715. [Google Scholar] [CrossRef]
- Spaan, A.N.; van Strijp, J.A.G.; Torres, V.J. Leukocidins: Staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 2017, 15, 435–447. [Google Scholar] [CrossRef]
- Yamada, T.; Tochimaru, N.; Nakasuji, S.; Hata, E.; Kobayashi, H.; Eguchi, M.; Kaneko, J.; Kamio, Y.; Kaidoh, T.; Takeuchi, S. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM–lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 2005, 110, 97–103. [Google Scholar] [CrossRef]
- Haveri, M.; Roslöf, A.; Rantala, L.; Pyörälä, S. Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J. Appl. Microbiol. 2007, 103, 993–1000. [Google Scholar] [CrossRef]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular Characterization of Staphylococcus aureus Isolated from Bovine Mastitis and Close Human Contacts in South African Dairy Herds: Genetic Diversity and Inter-Species Host Transmission. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Vrieling, M.; Boerhout, E.M.; van Wigcheren, G.F.; Koymans, K.J.; Mols-Vorstermans, T.G.; de Haas, C.J.C.; Aerts, P.C.; Daemen, I.J.J.M.; van Kessel, K.P.M.; Koets, A.P.; et al. LukMF′ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci. Rep. 2016, 6, 37759. [Google Scholar] [CrossRef]
- Ren, Q.; Liao, G.; Wu, Z.; Lv, J.; Chen, W. Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J. Dairy Sci. 2020, 103, 3368–3380. [Google Scholar] [CrossRef] [Green Version]
- Dhanawade, N.B.; Kalorey, D.R.; Srinivasan, R.; Barbuddhe, S.B.; Kurkure, N.V. Detection of intercellular adhesion genes and biofilm production in Staphylococcus aureus isolated from bovine subclinical mastitis. Vet. Res. Commun. 2010, 34, 81–89. [Google Scholar] [CrossRef]
- Li, T.; Lu, H.; Wang, X.; Gao, Q.; Dai, Y.; Shang, J.; Li, M. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front. Cell. Infect. Microbiol. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Bai, Y.; Xu, J.; Carter, M.Q.; Shi, C.; Shi, X. Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 2015, 195, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Liu, Q.; Gao, Q.; Lu, H.; Meng, H.; Xie, Y.; Huang, Q.; Ma, X.; Wang, H.; et al. Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mechesso, A.F.; Kim, S.J.; Park, H.S.; Choi, J.H.; Song, H.J.; Kim, M.H.; Lim, S.K.; Yoon, S.S.; Moon, D.C. Short communication: First detection of Panton-Valentine leukocidin–positive methicillin-resistant Staphylococcus aureus ST30 in raw milk taken from dairy cows with mastitis in South Korea. J. Dairy Sci. 2021, 104, 969–976. [Google Scholar] [CrossRef]
- Vaughn, J.M.; Abdi, R.D.; Gillespie, B.E.; Kerro Dego, O. Genetic diversity and virulence characteristics of Staphylococcus aureus isolates from cases of bovine mastitis. Microb. Pathog. 2020, 144, 104171. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Park, Y.K.; Koo, H.C.; Park, Y.H. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. J. Vet. Sci. 2010, 11, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Viçosa, G.N.; Le Loir, A.; Le Loir, Y.; de Carvalho, A.F.; Nero, L.A. egc characterization of enterotoxigenic Staphylococcus aureus isolates obtained from raw milk and cheese. Int. J. Food Microbiol. 2013, 165, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]


| Target | Sequence (5′ → 3′) | Size (bp) | References |
|---|---|---|---|
| nuc | F: GCGATTGATGGTGATACGGTT | 279 | [12] |
| R: AGCCAAGCCTTGACGAACTAAAGC | |||
| sea | F: GAAAAAAGTCTGAATTGCAGGGAACA | 560 | [13] |
| R: CAAATAAATCGTAATTAACCGAAGGTTC | |||
| seb | F:ATTCTATTAAGGACACTAAGTTAGGGA | 404 | [13] |
| R: ATCCCGTTTCATAAGGCGAGT | |||
| sec | F:CTTGTATGTATGGAGGAATAACAAAACATG | 275 | [13] |
| R: CATATCATACCAAAAAGTATTGCCGT | |||
| sed | F:GAATTAAGTAGTACCGCGCTAAATAATATG | 492 | [13] |
| R: GCTGTATTTTTCCTCCGAGAGT | |||
| see | F: CAAAGAAATGCTTTAAGCAATCTTAGGC | 482 | [13] |
| R: CACCTTACCGCCAAAGCTG | |||
| seg | F:TCTCCACCTGTTGAAGG | 323 | [13] |
| R: AAGTGATTGTCTATTGTCG | |||
| seh | F: CAATCACATCATATGCGAAAGCAG | 376 | [13] |
| R: CATCTACCCAAACATTAGCACC | |||
| sei | F: GTACCGTTGAAAATTCAG | 461 | [13] |
| R: AGGCAGTCCATCTCCTG | |||
| sej | F: TCAGAACTGTTGTTCCGCTAG | 138 | [13] |
| R: GAATTTTACCAYCAAAGGTAC | |||
| tst | F: CTGGTATAGTAGTGGGTCTG | 271 | [14] |
| R: AGGTAGTTCTATTGGAGTAGG | |||
| hla | F: CTGATTACTATCCAAGAAATTCGATTG | 209 | [15] |
| R: CTTTCCAGCCTACTTTTTTATCAGT | |||
| hlb | F: GTGCACTTACTGACAATAGTGC | 309 | [15] |
| R: GTTGATGAGTAGCTACCTTCAGT | |||
| lukS/F-PV | F: ATCATTAGGTAAAATGTCTGGACATGATCCA | 433 | [15] |
| R: GCATCAASTGTATTGGATAGCAAAAGC | |||
| lukED | F: TGAAAAAGGTTCAAAGTTGATACGAG | 269 | [16] |
| R: TGTATTCGATAGCAAAAGCAGTGCA | |||
| lukM | F: TGGATGTTACCTATGCAACCTAC | 780 | [15] |
| R: GTTCGTTTCCATATAATGAATCACTAC | |||
| fnbA | F: GTGAAGTTTTAGAAGGTGGAAAGATTAG | 643 | [17] |
| R: GCTCTTGTAAGACCATTTTTCTTCAC | |||
| fnbB | F: GTAACAGCTAATGGTCGAATTGATACT | 524 | [17] |
| R: CAAGTTCGATAGGAGTACTATGTTC | |||
| clfA | F: ATTGGCGTGGCTTCAGTGCT | 292 | [18] |
| R: CGTTTCTTCCGTAGTTGCATTTG | |||
| clfB | F: ACATCAGTAATAGTAGGGGGCAAC | 205 | [18] |
| R: TTCGCACTGTTTGTGTTTGCAC | |||
| icaA | F: CCTAACTAACGAAAGGTAG | 1315 | [19] |
| R: AAGATATAGCGATAAGTGC | |||
| icaD | F: AAACGTAAGAGAGGTGG | 381 | [19] |
| R: GGCAATATGATCAAGATAC |
| Virulence Gene Patterns | No. (%) of Isolates a | Factory (No. of Isolates) |
|---|---|---|
| hla, hlb, clfA, lukED | 2 (5.0) | A2 (1), B1 (1) |
| hla, hlb, clfB, lukED | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, clfB | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, fnbB, clfB | 1 (2.5) | A2 (1) |
| hla, hlb, fnbA, clfA, lukED | 6 (15.0) | A2 (6) |
| hla, hlb, fnbA, clfB, lukED | 4 (10.0) | A1 (3), A2 (1) |
| hla, hlb, fnbA, fnbB, lukED | 1 (2.5) | B2 (1) |
| hla, hlb, fnbA, clfA, icaD, lukED | 2 (5.0) | A2 (1), B1 (1) |
| hla, hlb, fnbA, clfB, icaA, lukED | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, clfB, icaD, lukED | 3 (7.5) | A1 (2), A2 (1) |
| hla, hlb, fnbA, fnbB, clfA, lukED | 6 (15.0) | A1 (3), B1 (1), C1 (2) |
| hla, hlb, fnbA, fnbB, icaD, lukED | 4 (10.0) | B2 (4) |
| hla, hlb, fnbA, clfB, icaA, icaD, lukED | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, fnbB, clfA, icaA, lukED | 2 (5.0) | A1 (2) |
| hla, hlb, fnbA, fnbB, icaA, icaD, lukED | 1 (2.5) | B2 (1) |
| hla, hlb, fnbA, fnbB, clfA, clfB, lukED | 1 (2.5) | A2 (1) |
| hla, hlb, fnbA, fnbB, clfB, icaD, lukED | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, fnbB, clfA, icaA, icaD, lukED | 1 (2.5) | A1 (1) |
| hla, hlb, fnbA, fnbB, clfB, icaD, lukED, tsst-1 | 1 (2.5) | A1 (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.-R.; Lee, Y.J. Characterization of Virulence Factors in Enterotoxin-Producing Staphylococcus aureus from Bulk Tank Milk. Animals 2022, 12, 301. https://doi.org/10.3390/ani12030301
Jung H-R, Lee YJ. Characterization of Virulence Factors in Enterotoxin-Producing Staphylococcus aureus from Bulk Tank Milk. Animals. 2022; 12(3):301. https://doi.org/10.3390/ani12030301
Chicago/Turabian StyleJung, Hye-Ri, and Young Ju Lee. 2022. "Characterization of Virulence Factors in Enterotoxin-Producing Staphylococcus aureus from Bulk Tank Milk" Animals 12, no. 3: 301. https://doi.org/10.3390/ani12030301
APA StyleJung, H.-R., & Lee, Y. J. (2022). Characterization of Virulence Factors in Enterotoxin-Producing Staphylococcus aureus from Bulk Tank Milk. Animals, 12(3), 301. https://doi.org/10.3390/ani12030301






