The Effect of Using Organic or Conventional Sires on Genetic Gain in Organic Pigs: A Simulation Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
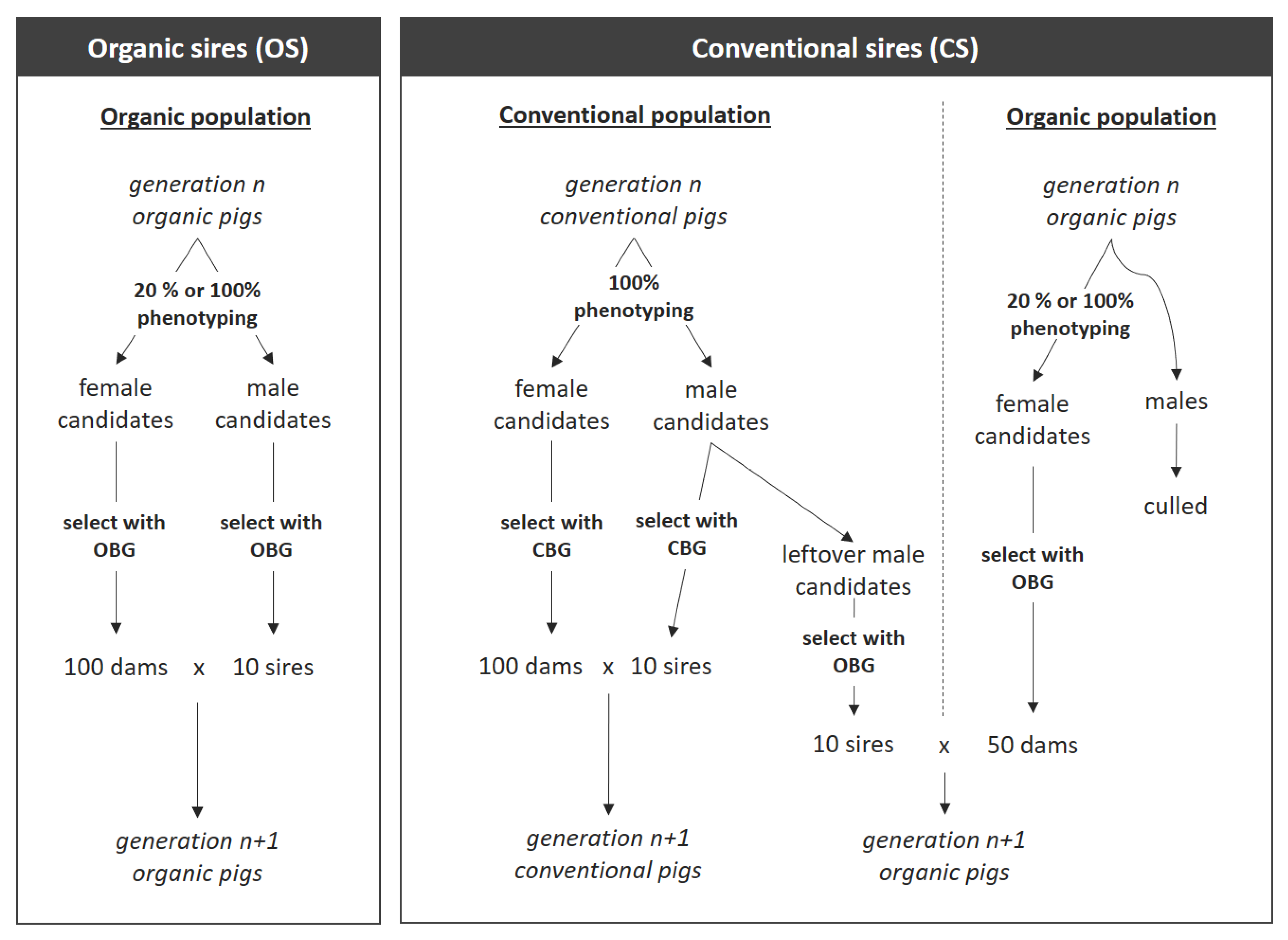
2.2. Overall Structure of Simulated Breeding Programs
2.2.1. Original Population of Sires
2.2.2. Breeding Goals
2.2.3. Traits
2.2.4. Phenotyping Intensity
2.3. Simulation Design
2.3.1. Population Structure
2.3.2. Selection of Dams and Sires
2.3.3. Estimation of Breeding Values
2.4. Data Analysis
3. Results
3.1. Genetic Correlations between Breeding Goals
3.2. Total Annual Genetic Gain and Rate of Inbreeding
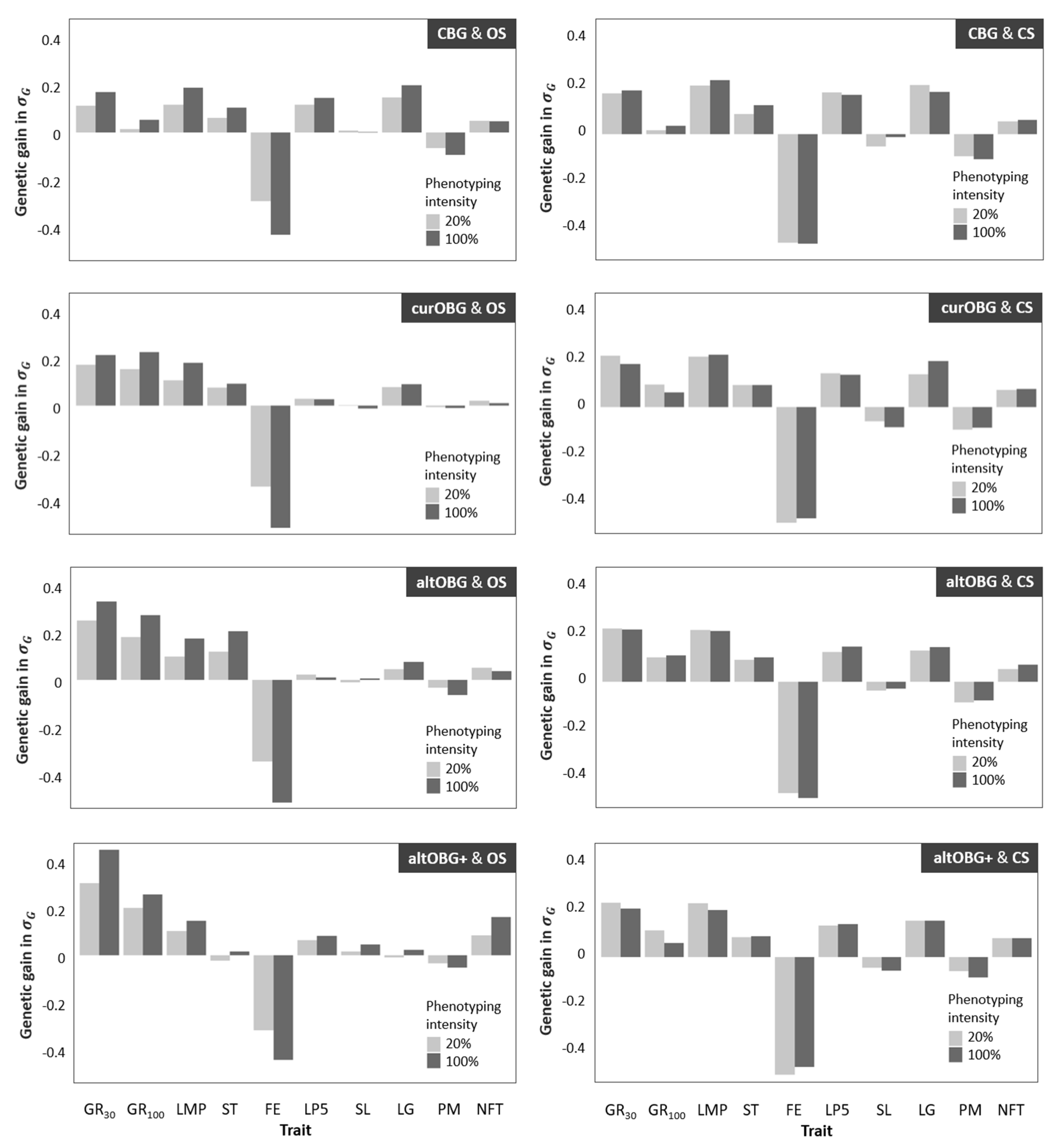
3.3. Genetic Gain for Individual Traits
4. Discussion
4.1. Conventional versus Organic Sires
4.2. Genetic Gain in Individual Traits
4.3. GxE Interactions
4.4. Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eurostat. Organic Livestock. Available online: https://ec.europa.eu/eurostat/databrowser/view/org_lstspec/default/table?lang=en (accessed on 3 May 2021).
- European Union. Organic Farming in the EU—A Fast Growing Sector; EU Agricultural Markets Briefs. No. 13; 2019; Available online: https://ec.europa.eu/info/sites/default/files/food-farming-fisheries/farming/documents/market-brief-organic-farming-in-the-eu_mar2019_en.pdf (accessed on 1 July 2021).
- Serup, T. Robust Racer—i Økologiske Sohold. Available online: https://www.frilandsdyr.dk/wp-content/uploads/2019/03/rapport-om-robuste-racer.pdf (accessed on 14 September 2021).
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics; Pearson Education Limited: Essex, UK, 1996. [Google Scholar]
- Mulder, H.A.; Bijma, P. Effects of genotype × environment interaction on genetic gain in breeding programs. J. Anim. Sci. 2005, 83, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Nirea, K.G.; Meuwissen, T.H.E. Improving production efficiency in the presence of genotype by environment interactions in pig genomic selection breeding programmes. J. Anim. Breed. Genet. 2017, 134, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Wallenbeck, A.; Rydhmer, L.; Lundeheim, N. GxE interactions for growth and carcass leanness: Re-ranking of boars in organic and conventional pig production. Livest. Sci. 2009, 123, 154–160. [Google Scholar] [CrossRef]
- Sørensen, A.C. Subjective definition of traits and economic values for selection of organic sows in Denmark. In Proceedings of the European Federation of Animal Science, Warsaw, Poland, 31 August–4 September 2015. [Google Scholar]
- Wallenbeck, A.; Rydhmer, L.; Röcklinsberg, H.; Ljung, M.; Strandberg, E.; Ahlman, T. Preferences for pig breeding goals among organic and conventional farmers in Sweden. Org. Agric. 2016, 6, 171–182. [Google Scholar] [CrossRef]
- Schild, S.L.; Baxter, E.M.; Pedersen, L.J. A review of neonatal mortality in outdoor organic production and possibilities to increase piglet survival. Appl. Anim. Behav. Sci. 2020, 231, 105088. [Google Scholar] [CrossRef]
- Pedersen, L.J.; Schild, S.-L.A.; Bonde, M.; Serup, T. Er der brug for søer med ny genetik i dansk økologisk svineproduktion? Okol. Erhverv. 2017, 12, 32495. [Google Scholar]
- Hermansen, J.E.; Lund, V.; Thuen, E. DARCOF Report No. 2: Ecological Animal Husbandry in the Nordic Countries. In Proceedings of the NJF-seminar No. 303, Horsens, Denmark, 16–17 September 1999. [Google Scholar]
- Knol, E.; Nielsen, B.; Knap, P.W. Genomic selection in commercial pig breeding. Anim. Front. 2016, 6, 15–22. [Google Scholar] [CrossRef]
- Wray, N.R.; Thompson, R. Prediction of rates of inbreeding in selected populations. Genet. Res. 1990, 55, 41–54. [Google Scholar] [CrossRef]
- Tess, M.W.; Bennett, G.L.; Dickerson, G.E. Simulation of genetic changes in life cycle efficiency of pork production, II. Effects of components on efficiency. J. Anim. Sci. 1983, 56, 354–368. [Google Scholar] [CrossRef]
- Gourdine, J.L.; de Greef, K.H.; Rydhmer, L. Breeding for welfare in outdoor pig production: A simulation study. Livest. Sci. 2010, 132, 26–34. [Google Scholar] [CrossRef]
- Ceron-Rojas, J.J.; Crossa, J.; Arief, V.N.; Basford, K.; Rutkoski, J.; Jarquín, D.; Alvarado, G.; Beyene, Y.; Semagn, K.; DeLacy, I. A genomic selection index applied to simulated and real data. Gen. Genom. Genet. 2015, 5, 2155–2164. [Google Scholar] [CrossRef] [PubMed]
- Thomasen, J.R.; Liu, H.; Sørensen, A.C. Genotyping more cows increases genetic gain and reduces rate of true inbreeding in a dairy cattle breeding scheme using female reproductive technologies. J. Dairy Sci. 2020, 103, 597–606. [Google Scholar] [CrossRef]
- Laval, G.; Iannuccelli, N.; Legault, C.; Milan, D.; Groenen, M.A.; Giuffra, E.; Andersson, L.; Nissen, P.H.; Jørgensen, C.B.; Beeckmann, P.; et al. Genetic diversity of eleven European pig breeds. Genet. Sel. Evol. 2000, 32, 187–203. [Google Scholar] [CrossRef] [PubMed]
- SanCristobal, M.; Chevalet, C.; Haley, C.; Joosten, R.; Rattink, A.P.; Harlizius, B.; Groenen, M.; Amigues, Y.; Boscher, M.-Y.; Russell, G.; et al. Genetic diversity within and between European pig breeds using microsatellite markers. Anim. Genet. 2006, 37, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.L.; Newman, S. Breeding for profit: Synergism between genetic improvement and livestock production (a review). J. Anim. Sci. 1994, 72, 2178–2200. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Quimm, T.H.M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Pedersen, L.D.; ASørensen, C.; Henryon, M.; Ansari-Mahyari, S.; Berg, P. ADAM: A computer program to simulate selective breeding schemes for animals. Livest. Sci. 2009, 121, 343–344. [Google Scholar] [CrossRef]
- Eurostat. Pig Population. Available online: https://ec.europa.eu/eurostat/databrowser/view/apro_mt_lspig/default/table?lang=en (accessed on 3 May 2021).
- SEGES. Differentietet Avl af Grise—Via Danavl. Available online: https://www.landbrugsinfo.dk/-/media/landbrugsinfo/public/0/0/9/diff_avl_af_oeko_grise_via_danavl.pdf (accessed on 10 November 2021).
- Lund, M.S.; Puonti, M.; Rydhmer, L.; Jensen, J. Relationship between litter size and perinatal and pre-weaning survival in pigs. Animal Science. Anim. Sci. 2002, 74, 217–222. [Google Scholar] [CrossRef]
- Su, G.; Lund, M.S.; Sorensen, D. Selection for litter size at day five to improve litter size at weaning and piglet survival rate. J. Anim. Sci. 2007, 85, 1385–1392. [Google Scholar] [CrossRef]
- Do, D.N.; Strathe, A.B.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genetic parameters for different measures of feed efficiency and related traits in boars of three pig breeds. J. Anim. Sci. 2013, 91, 4069–4079. [Google Scholar] [CrossRef]
- Holm, B.; Bakken, M.; Klemetsdal, G.; Vangen, O. Genetic correlations between reproduction and production traits in swine. J. Anim. Sci. 2004, 82, 3458–3464. [Google Scholar] [CrossRef]
- Andonov, S.; VVukovic, V.; Uzunov, A. Genetic Parameters for Reproductive Traits And Number of Teats in Pigs. In Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, 1–6 August 2010. [Google Scholar]
- Lundeheim, N.; Chalkias, H.; Rydhmer, L. Genetic analysis of teat number and litter traits in pigs. ACTA Sect. A—Anim. Sci. 2013, 63, 121–125. [Google Scholar] [CrossRef]
- Madsen, P.; Jensen, J. A User’s Guide to DMU; Version 6, Release 5.2; Aarhus University: Foulum, Denmark, 2013. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 1 July 2021).
- Strudsholm, K.; Hermansen, E. Performance and carcass quality of fully or partly outdoor reared pigs in organic production. Livest. Prod. Sci. 2005, 96, 261–268. [Google Scholar] [CrossRef]
- Slagboom, M.; Hjortø, L.; Sørensen, A.C.; Mulder, H.A.; Thomasen, J.R.; Kargo, M. Possibilities for a specific breeding program for organic dairy production. J. Dairy Sci. 2020, 103, 6332–6345. [Google Scholar] [CrossRef]
- Milligan, B.N.; Fraser, D.; Kramer, D.L. Within-litter birth weight variation in the domestic pig and its relation to pre-weaning survival, weight gain, and variation in weaning weights. Livest. Prod. Sci. 2002, 76, 181–191. [Google Scholar] [CrossRef]
- Quiniou, N.; Dagorn, J.; Gaudré, D. Variation of piglets’ birth weight and consequences on subsequent performance. Livest. Prod. Sci. 2002, 78, 63–70. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Kuhn, G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis. J. Amim. Sci. 2006, 84, E113–E123. [Google Scholar] [CrossRef]
- Town, S.C.; Putman, C.T.; Turchinsky, N.J.; Dixon, W.T.; Foxcroft, G.R. Number of conceptuses in utero affects porcine fetal muscle development. Reproduction 2004, 128, 443–454. [Google Scholar] [CrossRef]
- Högberg, A.; Rydhmer, L. A Genetic Study of Piglet Growth and Survival. Acta Agric. Scand. Sect. A Anim. Sci. 2000, 50, 300–330. [Google Scholar] [CrossRef]
- Damgaard, L.H.; Rydhmer, L.; Løvendahl, P.; Grandinson, K. Genetic parameters for within-litter variation in piglet birth weight and change in within-litter variation during suckling. J. Anim. Sci. 2003, 81, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Gaines, A.M. Herd Management Factors That Influence Whole Herd Feed Efficiency. Feed Efficiency in Swine; Wageningen Academic Publishers: Wageningen, The Netherlands, 2012; pp. 26–28. [Google Scholar]
- Leite, N.G.; Knol, E.F.; Garcia, A.L.; Lopes, M.S.; Zak, L.; Tsuruta, S.; Silva, F.F.; Lourenco, D. Investigating pig survival in different production phases using genomic models. J. Anim. Sci. 2021, 99, skab217. [Google Scholar] [CrossRef]
- Nielsen, H.M.; Olesen, I.; Navrud, S.; Kolstad, K.; Amer, P. How to Consider the Value of Farm Animals in Breeding Goals. A Review of Current Status and Future Challenges. J. Agric. Environ. Ethics 2011, 24, 309–330. [Google Scholar] [CrossRef]
- IFOAM. Principles of Organic Agriculture. Available online: https://www.ifoam.bio/en/organic-landmarks/principles-organic-agriculture (accessed on 25 November 2021).
- Merks, J.W.M. Genotype × Environment Interactions in Pig Breeding Programmes. VI. Genetic Relations between Performances in Central Test, On-farm Test and Commercial Fattening. Livest. Prod. Sci. 1989, 22, 325–339. [Google Scholar] [CrossRef]
- Brandt, H.; Werner, D.N.; Baulain, U.; Brade, W.; Weissmann, F. Genotype–environment interactions for growth and carcass traits in different pig breeds kept under conventional and organic production systems. Animal 2010, 4, 535–544. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brascamp, E.W.; Merks, J.W.M.; Wilmink, J.B.M. Genotype environment interaction in pig breeding programmes: Methods of estimation and relevance of the estimates. Livest. Prod. Sci. 1985, 13, 135–146. [Google Scholar] [CrossRef]
- Mulder, H.A.; Bijma, P. Benefits of Cooperation Between Breeding Programs in the Presence of Genotype by Environment Interaction. J. Dairy Sci. 2006, 89, 1727–1739. [Google Scholar] [CrossRef]
- Mulder, H.A.; Veerkamp, R.F.; Ducro, B.J.; van Arendonk, J.A.M.; Bijma, P. Optimization of Dairy Cattle Breeding Programs for Different Environments with Genotype by Environment Interaction. J. Dairy Sci. 2006, 5, 1740–1752. [Google Scholar] [CrossRef]
| Traits 1 | GR30 | GR100 | LMP | ST | FE | LP5 | SL | LG | PM | NFT |
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic parameters 2 | ||||||||||
| GR30 (g/day) | 0.29 | 0.46 | −0.04 | 0.00 | −0.20 | −0.05 | 0.00 | 0.00 | 0.00 | 0.19 |
| GR100 (g/day) | 0.06 | 0.33 | −0.20 | 0.00 | −0.30 | −0.15 | 0.00 | −0.25 | 0.05 | 0.00 |
| LMP (%) | 0.05 | 0.04 | 0.44 | 0.00 | −0.34 | 0.05 | 0.00 | −0.11 | 0.05 | 0.00 |
| ST (Points) | 0.00 | 0.00 | 0.00 | 0.17 | 0.00 | −0.10 | 0.00 | 0.13 | −0.15 | 0.00 |
| FE (FE/kg gain) | −0.04 | −0.53 | −0.07 | 0.00 | 0.32 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| LP5 (N/litter) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.06 | 0.00 | 0.26 | −0.40 | 0.13 |
| SL (kg) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.00 | 0.00 |
| LG (%) | 0.00 | 0.09 | 0.00 | 0.05 | 0.00 | 0.00 | 0.00 | 0.17 | 0.00 | 0.00 |
| PM (%) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.04 | 0.00 |
| NFT (Number) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.31 |
| Variances 2 | ||||||||||
| 185.000 | 1536.000 | 0.275 | 0.100 | 0.006 | 0.900 | 0.600 | 0.028 | 0.120 | 0.035 | |
| 637.931 | 4654.545 | 0.625 | 0.588 | 0.019 | 15.000 | 2.000 | 0.165 | 3.000 | 0.113 | |
| 452.931 | 3118.545 | 0.350 | 0.488 | 0.013 | 14.100 | 1.400 | 0.137 | 2.880 | 0.078 | |
| Economic values (€) 3 | ||||||||||
| CBG | 0.015 | 0.017 | 1.293 | 1.667 | −19.600 | 2.613 | −0.680 | 11.333 | 0 | 0 |
| curOBG | 0.012 | 0.029 | 1.533 | 1.667 | −29.333 | 0.693 | −1.747 | 11.333 | 0 | 0 |
| altOBG | 0.068 | 0.019 | 1.898 | 4.723 | −31.412 | 1.228 | −0.283 | 3.177 | 0 | 0 |
| altOBG+ | 0.113 | 0.013 | 1.029 | 0 | −23.659 | 1.631 | 0 | 0 | −2.232 | 3.753 |
| Breeding Goal | CBG | curOBG | altOBG | altOBG+ |
|---|---|---|---|---|
| Conventional (CBG) | - | 0.83 | 0.76 | 0.72 |
| Current organic (curOBG) | - | - | 0.87 | 0.69 |
| Alternative organic (farmer preferences) (altOBG) | - | - | - | 0.83 |
| Alternative organic + (additional traits) (altOBG+) | - | - | - | - |
| Sire Original Population | Phenotype Intensity | Breeding Goal 1 | Annual Genetic Gain for Individual Traits 2 | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GR30 | GR100 | LMP | ST | FE | LP5 | SL | LG | PM | NFT | ∆G 4 | ∆F 3 | ∆G/∆F 5 | |||
| OS | 20% | curOBG | 0.03 | 0.18 | 0.09 | 0.04 | 0.79 | 0.02 | 0.00 | 0.15 | 0.00 | 0.00 | 1.28 | 2.4 | 0.27 |
| altOBG | 0.04 | 0.21 | 0.08 | 0.06 | 0.80 | 0.01 | 0.01 | 0.09 | 0.00 | 0.00 | 1.31 | 2.4 | 0.27 | ||
| altOBG+ | 0.05 | 0.23 | 0.08 | −0.01 | 0.73 | 0.04 | −0.02 | −0.02 | 0.00 | 0.00 | 1.08 | 2.5 | 0.22 | ||
| 100% | curOBG | 0.04 | 0.26 | 0.15 | 0.05 | 1.19 | 0.02 | 0.02 | 0.17 | 0.00 | 0.00 | 1.89 | 4.4 | 0.21 | |
| altOBG | 0.05 | 0.32 | 0.14 | 0.11 | 1.19 | 0.01 | −0.01 | 0.14 | 0.00 | 0.00 | 1.96 | 3.7 | 0.26 | ||
| altOBG+ | 0.07 | 0.30 | 0.12 | 0.01 | 1.01 | 0.05 | −0.06 | 0.04 | 0.00 | 0.00 | 1.54 | 4.0 | 0.19 | ||
| CS | 20% | curOBG | 0.04 | 0.11 | 0.17 | 0.05 | 1.11 | 0.09 | 0.08 | 0.27 | 0.00 | 0.00 | 1.92 | 4.0 | 0.24 |
| altOBG | 0.04 | 0.12 | 0.18 | 0.05 | 1.08 | 0.08 | 0.05 | 0.25 | 0.00 | 0.00 | 1.84 | 4.0 | 0.23 | ||
| altOBG+ | 0.04 | 0.13 | 0.19 | 0.05 | 1.15 | 0.09 | 0.06 | 0.30 | 0.00 | 0.00 | 2.00 | 4.4 | 0.23 | ||
| 100% | curOBG | 0.03 | 0.07 | 0.18 | 0.05 | 1.07 | 0.09 | 0.12 | 0.37 | 0.00 | 0.00 | 1.98 | 3.9 | 0.25 | |
| altOBG | 0.04 | 0.13 | 0.17 | 0.05 | 1.13 | 0.1 | 0.04 | 0.28 | 0.00 | 0.00 | 1.94 | 4.0 | 0.24 | ||
| altOBG+ | 0.03 | 0.07 | 0.16 | 0.05 | 1.07 | 0.09 | 0.08 | 0.30 | 0.00 | 0.00 | 1.85 | 4.1 | 0.23 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaalberg, R.M.; Nielsen, H.M.; Sørensen, A.C.; Chu, T.T.; Jensen, J.; Villumsen, T.M. The Effect of Using Organic or Conventional Sires on Genetic Gain in Organic Pigs: A Simulation Study. Animals 2022, 12, 455. https://doi.org/10.3390/ani12040455
Zaalberg RM, Nielsen HM, Sørensen AC, Chu TT, Jensen J, Villumsen TM. The Effect of Using Organic or Conventional Sires on Genetic Gain in Organic Pigs: A Simulation Study. Animals. 2022; 12(4):455. https://doi.org/10.3390/ani12040455
Chicago/Turabian StyleZaalberg, Roos Marina, Hanne Marie Nielsen, Anders Christian Sørensen, Thinh T. Chu, Just Jensen, and Trine Michelle Villumsen. 2022. "The Effect of Using Organic or Conventional Sires on Genetic Gain in Organic Pigs: A Simulation Study" Animals 12, no. 4: 455. https://doi.org/10.3390/ani12040455
APA StyleZaalberg, R. M., Nielsen, H. M., Sørensen, A. C., Chu, T. T., Jensen, J., & Villumsen, T. M. (2022). The Effect of Using Organic or Conventional Sires on Genetic Gain in Organic Pigs: A Simulation Study. Animals, 12(4), 455. https://doi.org/10.3390/ani12040455






