A Field-Deployable Insulated Isothermal PCR (iiPCR) for the Global Surveillance of Toxoplasma gondii Infection in Cetaceans
Abstract
:Simple Summary
Abstract
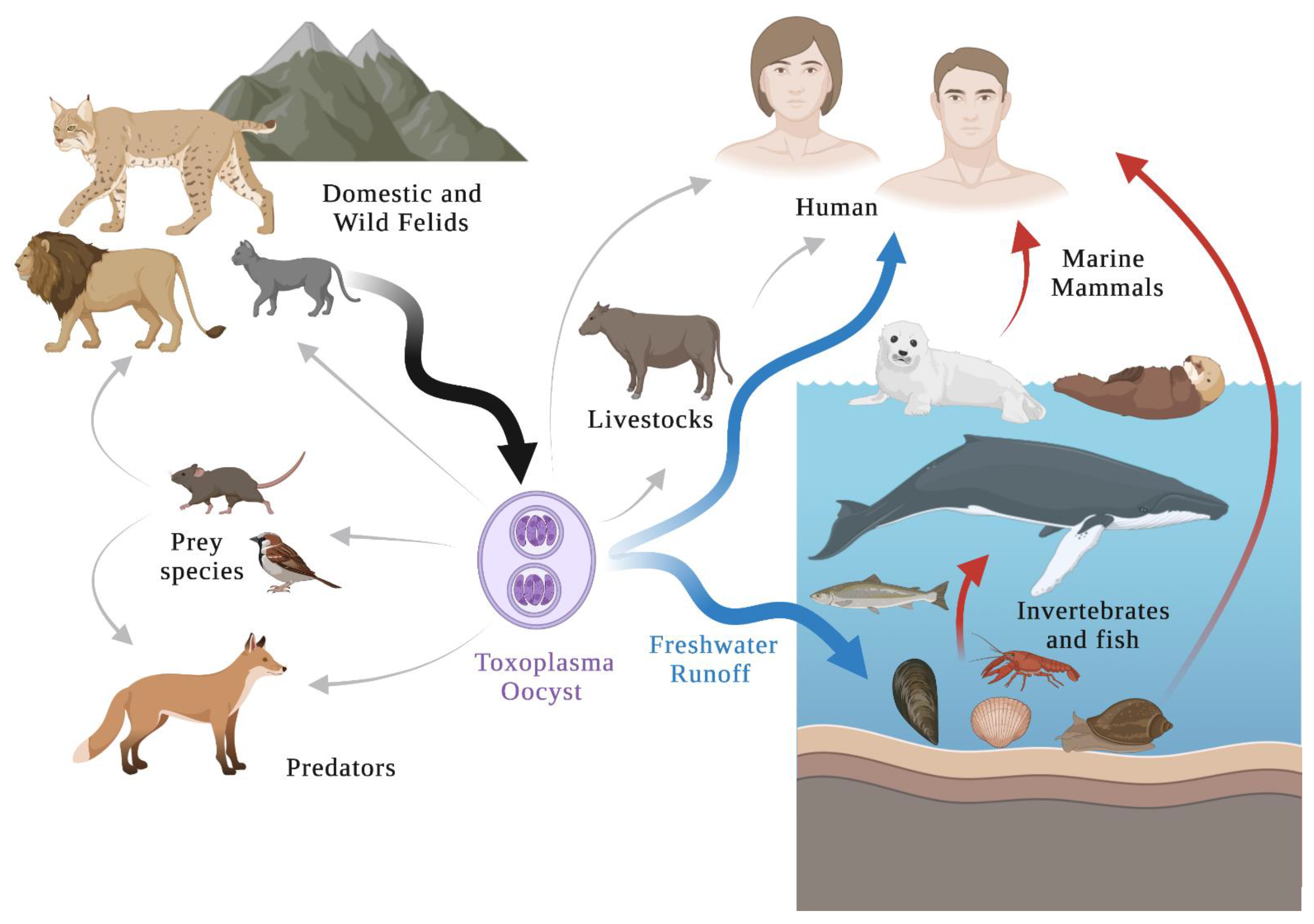
1. Introduction
2. Materials and Methods
2.1. Ethic Statement
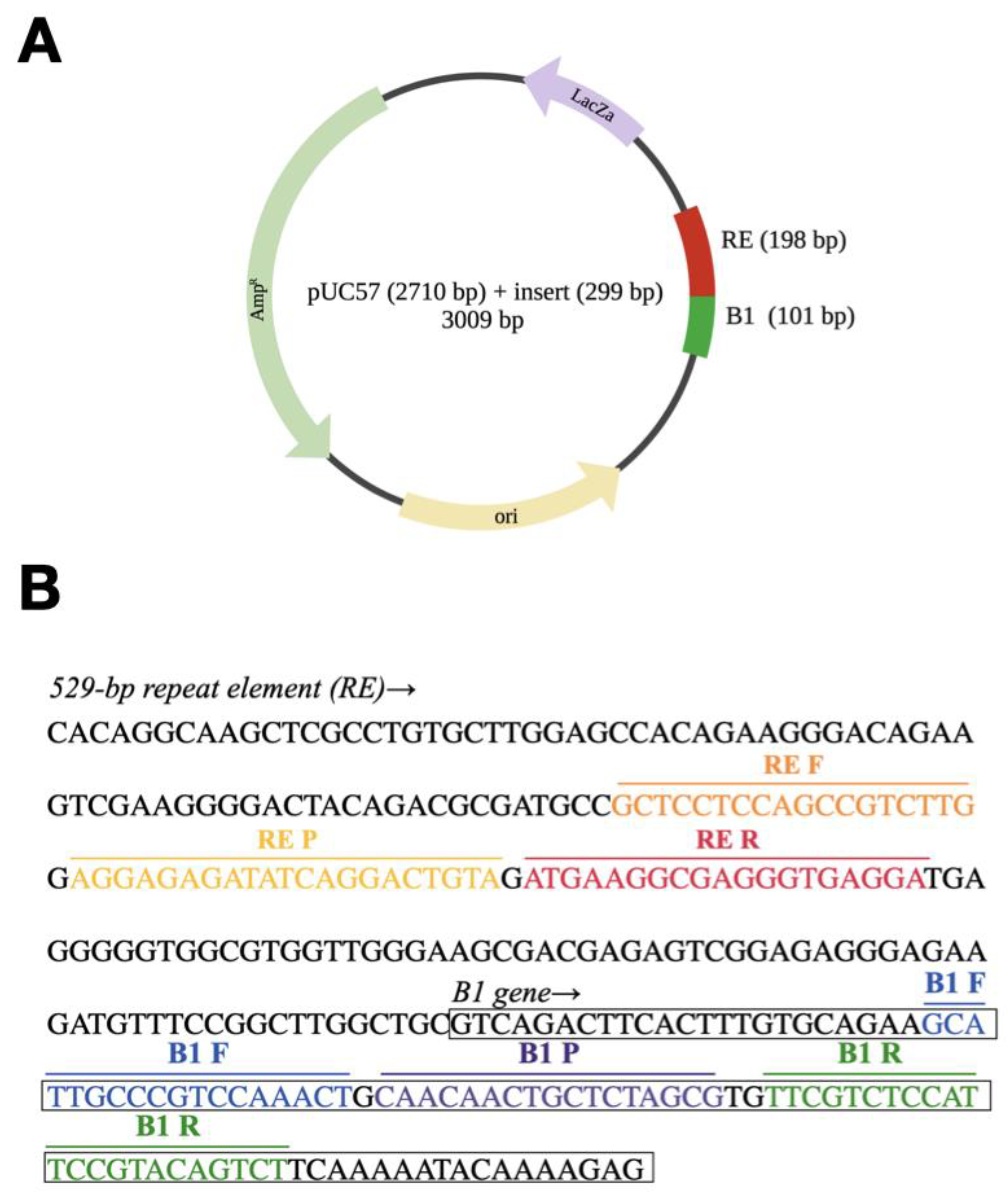
2.2. Synthetic Spike-in Standard
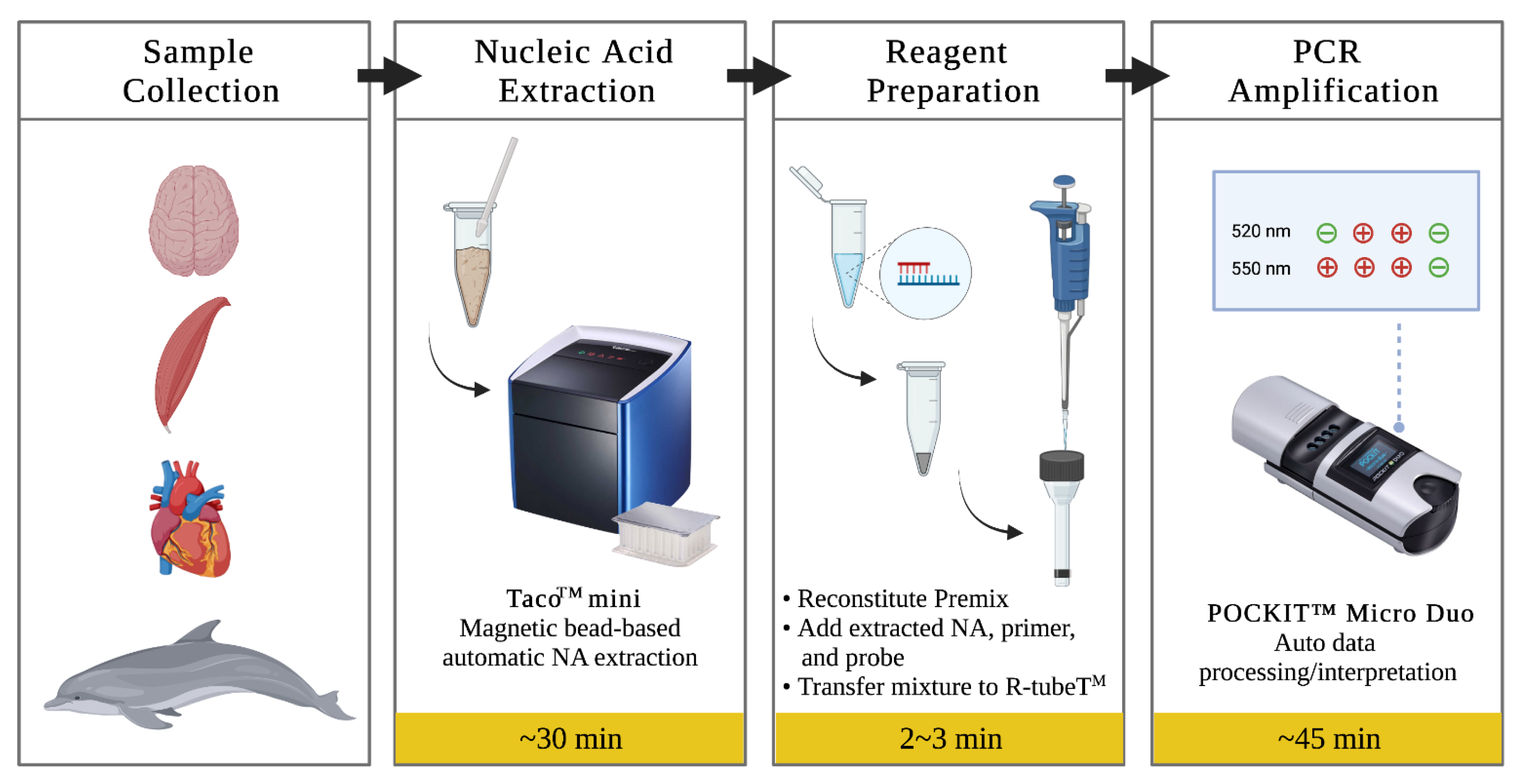
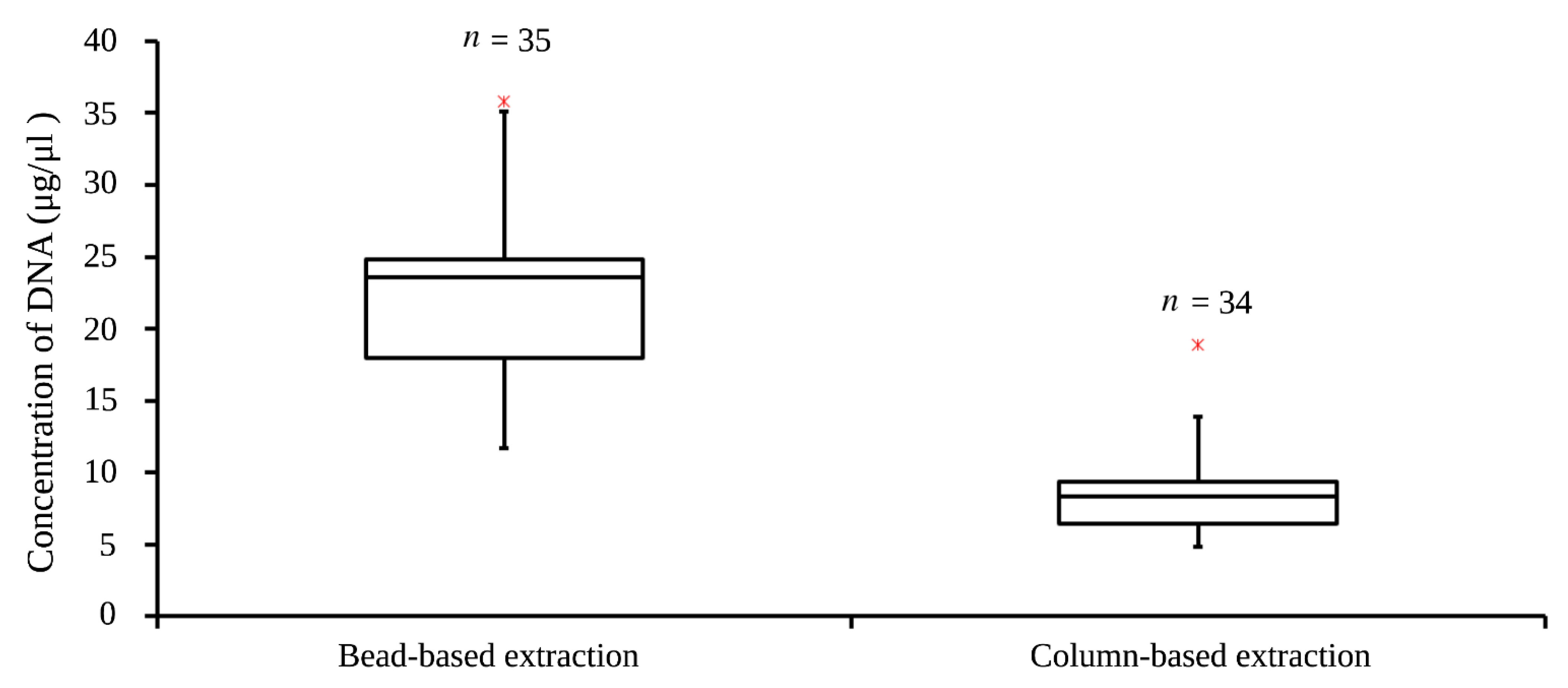
2.3. DNA Extraction
2.4. Insulated Isothermal PCR
2.5. Real-Time PCR
2.6. Statistics
3. Results
3.1. DNA Concentration
3.2. Analytical Specificity
3.3. Analytical Sensitivity
3.4. Sensitivity in Singleplex iiPCR
3.5. Sensitivity in Duplex iiPCR
3.6. Performance Evaluation of the B1/B2M iiPCR
3.6.1. Cerebrum Tissue
3.6.2. Cerebellum Tissue
3.6.3. Muscle Tissue
3.6.4. Myocardium Tissue
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dubey, J.P.; Jones, J.L. Toxoplasma gondii infection in humans and animals in the United States. Int. J. Parasitol. 2008, 38, 1257–1278. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, W.M.; Dunachie, J.F.; Siim, J.C.; Work, K. Life cycle of Toxoplasma gondii. Br. Med. J. 1969, 4, 806. [Google Scholar] [CrossRef] [Green Version]
- Miller, N.L.; Frenkel, J.K.; Dubey, J.P. Oral Infections with Toxoplasma Cysts and Oocysts in Felines, Other Mammals, and in Birds. J. Parasitol. 1972, 58, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P. The history of Toxoplasma gondii—The first 100 years. J. Eukaryot. Microbiol. 2008, 55, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P. Toxoplasmosis—A waterborne zoonosis. Vet. Parasitol. 2004, 126, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, A.; Blanchard, N.; Besteiro, S. The bradyzoite: A key developmental stage for the persistence and pathogenesis of toxoplasmosis. Pathogens 2020, 9, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, J.P.; Frenkel, J.K. Cyst-Induced Toxoplasmosis in Cats. J. Protozool. 1972, 19, 155–177. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; Frenkel, J.K. Experimental Toxoplasma Infection in Mice with Strains Producing Oocysts. J. Parasitol. 1973, 59, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Davidson, M.G.; Rottman, J.B.; English, R.V.; Lappin, M.R.; Tompkins, M.B. Feline immunodeficiency virus predisposes cats to acute generalized toxoplasmosis. Am. J. Pathol. 1993, 143, 1486–1497. [Google Scholar] [PubMed]
- Berdoy, M.; Webster, J.P.; Mcdonald, D.W. Fatal attraction in rats infected with Toxoplasma gondii. Proc. R. Soc. B Biol. Sci. 2000, 267, 1591–1594. [Google Scholar] [CrossRef] [Green Version]
- Vyas, A.; Kim, S.K.; Giacomini, N.; Boothroyd, J.C.; Sapolsky, R.M. Behavioral changes induced by Toxoplasma infection of rodents are highly specific to aversion of cat odors. Proc. Natl. Acad. Sci. USA 2007, 104, 6442–6447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gering, E.; Laubach, Z.M.; Weber, P.S.D.; Soboll Hussey, G.; Lehmann, K.D.S.; Montgomery, T.M.; Turner, J.W.; Perng, W.; Pioon, M.O.; Holekamp, K.E.; et al. Toxoplasma gondii infections are associated with costly boldness toward felids in a wild host. Nat. Commun. 2021, 12, 3842. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Grigg, M.E.; Kreuder, C.; James, E.R.; Melli, A.C.; Crosbie, P.R.; Jessup, D.A.; Boothroyd, J.C.; Brownstein, D.; Conrad, P.A. An unusual genotype of Toxoplasma gondii is common in California sea otters (Enhydra lutris nereis) and is a cause of mortality. Int. J. Parasitol. 2004, 34, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Tenter, A.M.; Heckeroth, A.R.; Weiss, L.M. Toxoplasma gondii: From animals to humans. Int. J. Parasitol. 2000, 30, 1217–1258. [Google Scholar] [CrossRef] [Green Version]
- Abbas, I.E.; Villena, I.; Dubey, J.P. A review on toxoplasmosis in humans and animals from Egypt. Parasitology 2020, 147, 135–159. [Google Scholar] [CrossRef] [PubMed]
- Cenci-Goga, B.T.; Rossitto, P.V.; Sechi, P.; McCrindle, C.M.E.; Cullor, J.S. Toxoplasma in animals, food, and humans: An old parasite of new concern. Foodborne Pathog. Dis. 2011, 8, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, S.K.; Fitza, M.A.; Lerner, D.A.; Calhoun, D.M.; Beldon, M.A.; Chan, E.T.; Johnson, P.T.J. Risky business: Linking Toxoplasma gondii infection and entrepreneurship behaviours across individuals and countries. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, P.A.; Miller, M.A.; Kreuder, C.; James, E.R.; Mazet, J.; Dabritz, H.; Jessup, D.A.; Gulland, F.; Grigg, M.E. Transmission of Toxoplasma: Clues from the study of sea otters as sentinels of Toxoplasma gondii flow into the marine environment. Int. J. Parasitol. 2005, 35, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, K.; Conrad, P.A.; Mazet, J.A.K.; Wallender, W.W.; Miller, W.A.; Largier, J.L. Effect of estuarine wetland degradation on transport of Toxoplasma gondii surrogates from land to sea. Appl. Environ. Microbiol. 2010, 76, 6821–6828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanWormer, E.; Fritz, H.; Shapiro, K.; Mazet, J.A.K.; Conrad, P.A. Molecules to modeling: Toxoplasma gondii oocysts at the human–animal–environment interface. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 217–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, D.S.; Collins, M.V.; Mitchell, S.M.; Cole, R.A.; Flick, G.J.; Wetch, C.N.; Lindquist, A.; Dubey, J.P. Sporulation and Survival of Toxoplasma gondii Oocysts in Seawater. J. Eukaryot. Microbiol. 2003, 50, 687–688. [Google Scholar] [CrossRef]
- Wainwright, K.E.; Miller, M.A.; Melli, A.C.; Packham, A.E. Chemical inactivation of Toxoplasma gondii oocusts in water. Society 2007, 93, 925–931. [Google Scholar]
- Wainwright, K.E.; Lagunas-Solar, M.; Miller, M.A.; Barr, B.C.; Gardner, I.A.; Pina, C.; Melli, A.C.; Packham, A.E.; Zeng, N.; Truong, T.; et al. Physical inactivation of Toxoplasma gondii oocysts in water. Appl. Environ. Microbiol. 2007, 73, 5663–5666. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, K.; Bahia-Oliveira, L.; Dixon, B.; Dumètre, A.; de Wit, L.A.; VanWormer, E.; Villena, I. Environmental transmission of Toxoplasma gondii: Oocysts in water, soil and food. Food Waterborne Parasitol. 2019, 15, e00049. [Google Scholar] [CrossRef]
- Massie, G.N.; Ware, M.W.; Villegas, E.N.; Black, M.W. Uptake and transmission of Toxoplasma gondii oocysts by migratory, filter-feeding fish. Vet. Parasitol. 2010, 169, 296–303. [Google Scholar] [CrossRef]
- Ratcliffe, H.L.; Worth, C.B. Toxoplasmosis of captive wild birds and mammals. Am. J. Pathol. 1951, 27, 655–667. [Google Scholar]
- Miller, M.; Shapiro, K.; Murray, M.J.; Haulena, M.; Raverty, S. Protozoan Parasites of Marine Mammals. In CRC Handbook of Marine Mammal Medicine; Gulland, F.M.D., Dierauf, L.A., Whitman, K.L., Eds.; CRC Press: New York, NY, USA, 2018; ISBN 9781315144931. [Google Scholar]
- Bachand, N.; Ravel, A.; Leighton, P.; Stephen, C.; Ndao, M.; Avard, E.; Jenkins, E. Serological and molecular detection of Toxoplasma gondii in terrestrial and marine wildlife harvested for food in Nunavik, Canada. Parasites Vectors 2019, 12, 155. [Google Scholar] [CrossRef] [Green Version]
- Reiling, S.; Dixon, B. Toxoplasma gondii: How an Amazonian parasite became an Inuit health issue. Can. Commun. Dis. Rep. 2019, 45, 183–190. [Google Scholar] [CrossRef]
- Ducrocq, J.; Ndao, M.; Yansouni, C.P.; Proulx, J.F.; Mondor, M.; Hamel, D.; Lévesque, B.; De Serres, G.; Talbot, D. Epidemiology associated with the exposure to Toxoplasma gondii in Nunavik’s Inuit population using the 2017 Qanuilirpitaa cross-sectional health survey. Zoonoses Public Health 2021, 68, 803–814. [Google Scholar] [CrossRef]
- Sing-Ming, W.; Hung-Chan, W.; Yi-Ping, L.; Jiang-Ping, W.; Wei-Cheng, Y.; Chao-Nan, L.; Ming-Tang, C.; Ching-Dong, C.; Shinn-Shyong, T.; Tsung-Chou, C. A case report: Toxoplasmosis Encephalitis and Myocarditis in Finless porpoise (Neophocaena phocaenoides). In Proceedings of the Chinese Society of Veterinary Sciences and Taiwan Provincial Animal Husbandry and Veterinary Society 2013 Annual Spring Joint Academic Symposium, Taipei, Taiwan, 25 May 2013. [Google Scholar]
- Obusan, M.C.M.; Villanueva, R.M.D.; Siringan, M.A.T.; Rivera, W.L.; Aragones, L.V. Leptospira spp. And Toxoplasma gondii in stranded representatives of wild cetaceans in the Philippines. BMC Vet. Res. 2019, 15, 372. [Google Scholar] [CrossRef]
- Bigal, E.; Morick, D.; Scheinin, A.P.; Salant, H.; Berkowitz, A.; King, R.; Levy, Y.; Melero, M.; Sánchez-Vizcaíno, J.M.; Goffman, O.; et al. Detection of Toxoplasma gondii in three common bottlenose dolphins (Tursiops truncatus); A first description from the Eastern Mediterranean Sea. Vet. Parasitol. 2018, 258, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Bang-Yeh, L. Investigation of Ag Deposition/Distribution and Its Associations with Lesion Development and Infectious Diseases in Stranded Cetaceans. Master’s Thesis, National Taiwan University, Taipei, Taiwan, 2018. [Google Scholar]
- Roe, W.D.; Howe, L.; Baker, E.J.; Burrows, L.; Hunter, S.A. An atypical genotype of Toxoplasma gondii as a cause of mortality in Hector’s dolphins (Cephalorhynchus hectori). Vet. Parasitol. 2013, 192, 67–74. [Google Scholar] [CrossRef]
- Page-Karjian, A.; Lo, C.F.; Ritchie, B.; Harms, C.A.; Rotstein, D.S.; Han, S.; Hassan, S.M.; Lehner, A.F.; Buchweitz, J.P.; Thayer, V.G.; et al. Anthropogenic Contaminants and Histopathological Findings in Stranded Cetaceans in the Southeastern United States, 2012–2018. Front. Mar. Sci. 2020, 7, 630. [Google Scholar] [CrossRef]
- Sierra, E.; Fernández, A.; Felipe-Jiménez, I.; Zucca, D.; Díaz-Delgado, J.; Puig-Lozano, R.; Câmara, N.; Consoli, F.; Díaz-Santana, P.; Suárez-Santana, C.; et al. Histopathological Differential Diagnosis of Meningoencephalitis in Cetaceans: Morbillivirus, Herpesvirus, Toxoplasma gondii, Brucella sp., and Nasitrema sp. Front. Vet. Sci. 2020, 7, 650. [Google Scholar] [CrossRef] [PubMed]
- Grattarola, C.; Giorda, F.; Iulini, B.; Pintore, M.D.; Pautasso, A.; Zoppi, S.; Goria, M.; Romano, A.; Peletto, S.; Varello, K.; et al. Meningoencephalitis and Listeria monocytogenes, Toxoplasma gondii and Brucella spp. coinfection in a dolphin in Italy. Dis. Aquat. Organ. 2016, 118, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Paul, E. Emerging Diseases in Marine Mammals: From Dolphins to Manatees. Microbe 2009, III, 95–117. [Google Scholar] [CrossRef] [Green Version]
- Stewart, J.R.; Gast, R.J.; Fujioka, R.S.; Solo-Gabriele, H.M.; Meschke, J.S.; Amaral-Zettler, L.A.; Del Castillo, E.; Polz, M.F.; Collier, T.K.; Strom, M.S.; et al. The coastal environment and human health: Microbial indicators, pathogens, sentinels and reservoirs. Environ. Health A Glob. Access Sci. Source 2008, 7, S3. [Google Scholar] [CrossRef] [Green Version]
- Bossart, G.D. Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 2011, 48, 676–690. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, Z.D.; Huang, S.Y.; Zhu, X.Q. Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasites Vectors 2015, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Ajzenberg, D.; Lamaury, I.; Demar, M.; Vautrin, C.; Cabié, A.; Simon, S.; Nicolas, M.; Desbois-Nogard, N.; Boukhari, R.; Riahi, H.; et al. Performance Testing of PCR Assay in Blood Samples for the Diagnosis of Toxoplasmic Encephalitis in AIDS Patients from the French Departments of America and Genetic Diversity of Toxoplasma gondii: A Prospective and Multicentric Study. PLoS Negl. Trop. Dis. 2016, 10, e0004790. [Google Scholar] [CrossRef]
- Wahab, T.; Edvinsson, B.; Palm, D.; Lindh, J. Comparison of the AF146527 and B1 repeated elements, two real-time PCR targets used for detection of Toxoplasma gondii. J. Clin. Microbiol. 2010, 48, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.H.; Chou, L.S.; Chou, S.J.; Wang, J.H.; Stott, J.; Blanchard, M.; Jen, I.F.; Yang, Y.C. Selection of suitable reference genes for normalization of quantitative RT-PCR in peripheral blood samples of bottlenose dolphins (Tursiops truncatus). Sci. Rep. 2015, 5, 15425. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.-H.; Wang, J.-H.; Chou, S.-J.; Wu, Y.-H.; Li, T.-H.; Leu, M.-Y.; Chang, W.-B.; Yang, W.C. Selection of reference genes for RT-qPCR studies in blood of beluga whales (Delphinapterus leucas). PeerJ 2016, 4, e1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzariol, S.; Marcer, F.; Mignone, W.; Serracca, L.; Goria, M.; Marsili, L.; Di Guardo, G.; Casalone, C. Dolphin Morbillivirus and Toxoplasma gondii coinfection in a Mediterranean fin whale (Balaenoptera physalus). BMC Vet. Res. 2012, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Pretti, C.; Mancianti, F.; Nardoni, S.; Ariti, G.; Monni, G.; Bello, D.D.I.; Marsili, S.; Papini, R. Detection of Toxoplasma gondii infection in dolphins stranded along the Tuscan coast Italy. Rev. Med. Vet. 2010, 161, 428–431. [Google Scholar]
- Sotiriadou, I.; Karanis, P. Evaluation of loop-mediated isothermal amplification for detection of Toxoplasma gondii in water samples and comparative findings by polymerase chain reaction and immunofluorescence test (IFT). Diagn. Microbiol. Infect. Dis. 2008, 62, 357–365. [Google Scholar] [CrossRef]
- Wu, Y.D.; Xu, M.J.; Wang, Q.Q.; Zhou, C.X.; Wang, M.; Zhu, X.Q.; Zhou, D.H. Recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for detection of Toxoplasma gondii in the environment. Vet. Parasitol. 2017, 243, 199–203. [Google Scholar] [CrossRef]
- Schares, G.; Globokar Vrhovec, M.; Tuschy, M.; Joeres, M.; Bärwald, A.; Koudela, B.; Dubey, J.P.; Maksimov, P.; Conraths, F.J. A real-time quantitative polymerase chain reaction for the specific detection of Hammondia hammondi and its differentiation from Toxoplasma gondii. Parasites Vectors 2021, 14, 78. [Google Scholar] [CrossRef]
- Dubey, J.P.; Sreekumar, C. Redescription of Hammondia hammondi and its differentiation from Toxoplasma gondii. Int. J. Parasitol. 2003, 33, 1437–1453. [Google Scholar] [CrossRef]
- Lorenzi, H.; Khan, A.; Behnke, M.S.; Namasivayam, S.; Swapna, L.S.; Hadjithomas, M.; Karamycheva, S.; Pinney, D.; Brunk, B.P.; Ajioka, J.W.; et al. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun. 2016, 7, 10147. [Google Scholar] [CrossRef]
- Sreekumar, C.; Vianna, M.C.B.; Hill, D.E.; Miska, K.B.; Lindquist, A.; Dubey, J.P. Differential detection of Hammondia hammondi from Toxoplasma gondii using polymerase chain reaction. Parasitol. Int. 2005, 54, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Schares, G.; Ziller, M.; Herrmann, D.C.; Globokar, M.V.; Pantchev, N.; Conraths, F.J. Seasonality in the proportions of domestic cats shedding Toxoplasma gondii or Hammondia hammondi oocysts is associated with climatic factors. Int. J. Parasitol. 2016, 46, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Montazeri, M.; Mikaeili Galeh, T.; Moosazadeh, M.; Sarvi, S.; Dodangeh, S.; Javidnia, J.; Sharif, M.; Daryani, A. The global serological prevalence of Toxoplasma gondii in felids during the last five decades (1967-2017): A systematic review and meta-analysis. Parasites Vectors 2020, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Gao, Y.M.; Deng, Y.; Lamberton, P.H.L.; Lu, D.B. A systematic review and meta-analysis of the seroprevalence of Toxoplasma gondii in cats in mainland China. Parasites Vectors 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulland, F.M.D.; Lowenstine, L.J.; Lapointe, J.M.; Spraker, T.; King, D.P. Herpesvirus of Coastal Infection California in Stranded Pacific Harbor Seals of Coastal California. J. Wildl. Dis. 1997, 33, 450–458. [Google Scholar] [CrossRef]
- Schulman, F.Y.; Lipscomb, T.P.; Moffett, D.; Krafft, A.E.; Lichy, J.H.; Tsai, M.M.; Taubenberger, J.K.; Kennedy, S. Histologic, Immunohistochemical, and Polymerase Chain Reaction Studies of Bottlenose Dolphins from the 1987-1988 United States Atlantic Coast Epizootic. Vet. Pathol. 1997, 34, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, A.Y.; Reguzova, A.Y.; Rozanova, E.I.; Abramov, A.V.; Tumanov, Y.V.; Kuvshinova, I.N.; Shestopalov, A.M. Detection of specific antibodies to morbilliviruses, Brucella and Toxoplasma in the Black Sea dolphin Tursiops truncatus ponticus and the beluga whale Delphinapterus leucas from the Sea of Okhotsk in 2002-2007. Russ. J. Mar. Biol. 2009, 35, 494–497. [Google Scholar] [CrossRef]
- Shapiro, K. Climate and coastal habitat change: A recipe for a dirtier ocean. Mar. Pollut. Bull. 2012, 64, 1079–1080. [Google Scholar] [CrossRef] [PubMed]
- Burek, K.A.; Gulland, F.M.D.; O’Hara, T.M. Effects of climate change on arctic marine mammal health. Ecol. Appl. 2008, 18, 126–134. [Google Scholar] [CrossRef] [PubMed]


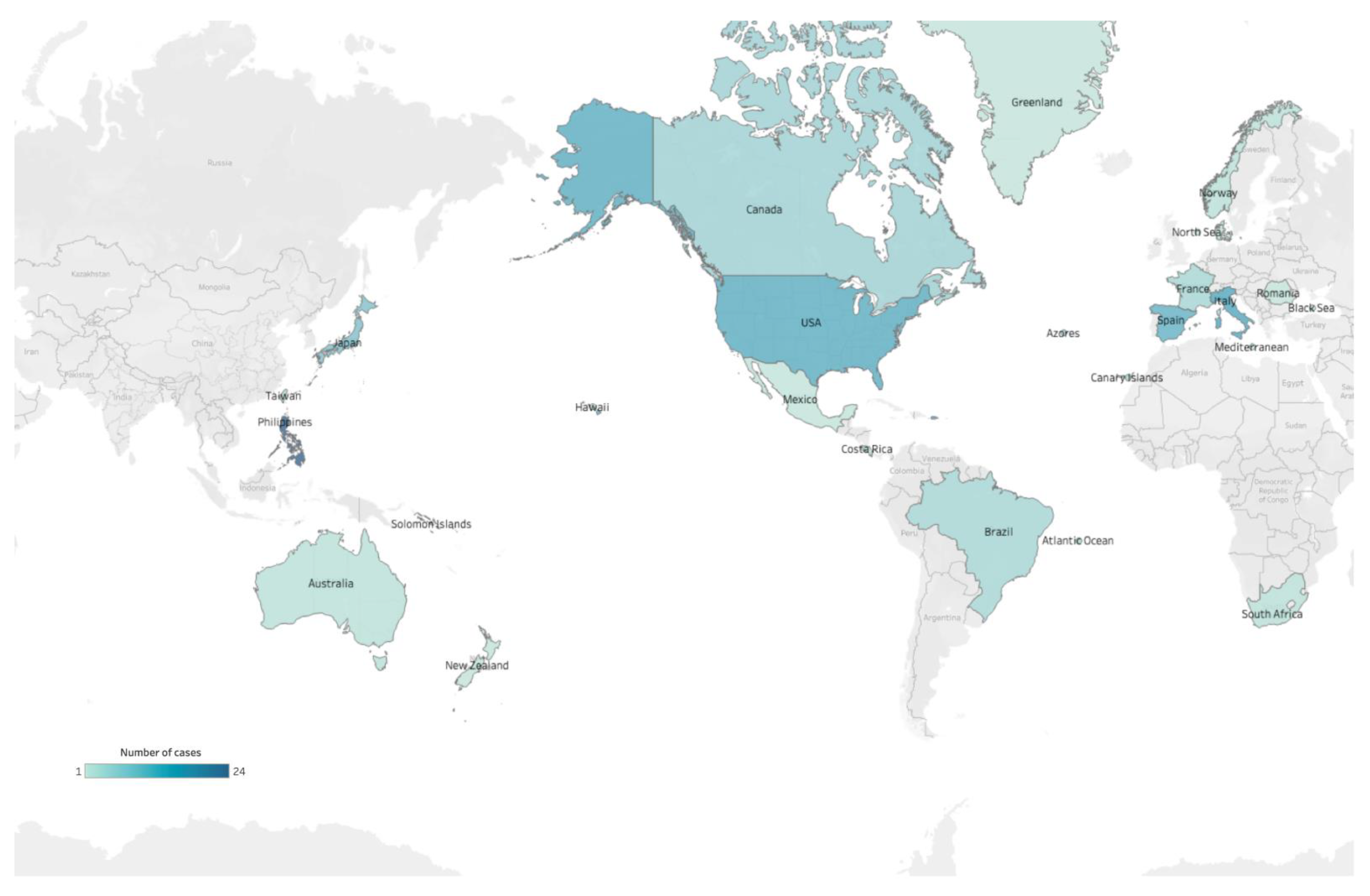

| Family | Genus | Species |
|---|---|---|
| Balaenopteridae | Balaenoptera | Blue whale (Balaenoptera musculus) |
| Balaenopteridae | Balaenoptera | Bryde’s whale (Balaenoptera edeni) |
| Balaenopteridae | Balaenoptera | Fin whale (Balaenoptera physalus) |
| Balaenopteridae | Balaenoptera | Minke whale (Balaeneoptera acutorostrata) |
| Balaenopteridae | Balaenoptera | Sei whale (Balaenoptera borealis) |
| Balaenopteridae | Megaptera | Humpback whale (Megaptera novaeangliae) |
| Delphinidae | Cephalorhynchus | Hector’s dolphin (Cephalorhynchus hectori) |
| Delphinidae | Delphinus | Common dolphin (Delphinus delphis) |
| Delphinidae | Feresa | Pygmy killer whale (Feresa attenuata) |
| Delphinidae | Globicephala | Pilot whale (Globicephala macrorhynchus) |
| Delphinidae | Globicephala | Pilot whale (Globicephala melaena) |
| Delphinidae | Grampus | Risso’s dolphin (Grampus griseus) |
| Delphinidae | Lagenodelphis | Fraser’s dolphin (Lagenodelphis hosei) |
| Delphinidae | Lagenorhynchus | White-sided dolphin (Lagenorynchus obliquidens) |
| Delphinidae | Orcinus | Killer whale (Orcinus orca) |
| Delphinidae | Peponocephala | Melon-headed whale (Peponocephala electra) |
| Delphinidae | Pseudorca | False killer whale (Pseudorca crassidens) |
| Delphinidae | Sotalia | Guiana dolphin (Sotalia guianensis) |
| Delphinidae | Sousa | Humpbacked dolphin (Sousa chinensis) |
| Delphinidae | Stenella | Pantropical spotted dolphin (Stenella attenuata) |
| Delphinidae | Stenella | Spinner dolphin (Stenella longirostris) |
| Delphinidae | Stenella | Spotted dolphin (Stenella frontalis) |
| Delphinidae | Stenella | Striped dolphin (Stenella coeruleoalba) |
| Delphinidae | Steno | Rough-toothed dolphin (Steno bredanensis) |
| Delphinidae | Tursiops | Bottlenose dolphin (Tursiops aduncus) |
| Delphinidae | Tursiops | Bottlenose dolphin (Tursiops truncatus) |
| Iniidae | Inia | Amazon River dolphin (Inia geoffrensis) |
| Kogiidae | Kogia | Pygmy sperm whale (Kogia breviceps) |
| Monodontidae | Delphinapterus | Beluga (Delphinapterus leucas) |
| Monodontidae | Monodon | Narwhal (Monodon nonoceros) |
| Phocoenidae | Neophocaena | Finless porpoise (Neophocaena phocaenoides) |
| Phocoenidae | Phocoena | Harbor porpoise (Phocoena phocoena) |
| Physeteridae | Physeter | Sperm whale (Physeter macrocephalus) |
| Ziphiidae | Mesoplodon | Beaked whale (Mesoplodon sp.) |
| Primers and Probes | Nucleotide Sequences (5′–3′) | Target Genes | Function | Amplicons | References |
|---|---|---|---|---|---|
| RE-R | 5′-TCCTCACCCTCGCCTTCAT-3′ | 529-bp repeat element | Unknown function | 60 bp | [45] |
| RE-F | 5′-GCTCCTCCAGCCGTCTTG-3′ | ||||
| RE-P | 5′-6-FAM-AGGAGAGATATCAGGACTGTA -MGB-NFQ -3 | ||||
| B1-R | 5′-AGACTGTACGGAATGGAGACGAA-3′ | B1 gene | cGMP dependent protein kinase activity | 61 bp | [44] |
| B1-F | 5′-GCATTGCCCGTCCAAACT-3′ | ||||
| B1-P | 5′-6-FAM–CAACAACTGCTCTAGCG–MGB-NFQ-3′ | ||||
| B2M-R | 5′- GCGTTGGGAGTGAACTCAG-3′ | B2M gene | β2-microglobulin | 78 bp | [46,47] |
| B2M-F | 5′-GGTGGAGCAATCAGACCTGT-3′ | ||||
| B2M-P | 5′-VIC-TCAGCAAGGACTGGTCTT-MGB-NFQ-3′ |
| Standard (Copy/μL) | RE qPCR No. Positive/ No. Tested | Rate (%) | RE iiPCR No. Positive/ No. Tested | Rate (%) | B1 qPCR No. Positive/ No. Tested | Rate (%) | B1 iiPCR No. Positive/ No. Tested | Rate (%) |
|---|---|---|---|---|---|---|---|---|
| 3 × 107 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 |
| 3 × 106 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 |
| 3 × 105 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 | 2/2 | 100 |
| 3 × 104 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 |
| 3 × 103 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 |
| 3 × 102 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 |
| 3 × 101 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 |
| 3 × 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 | 4/4 | 100 |
| 3 × 10−1 | 2/4 | 50 | 2/4 | 50 | 2/4 | 50 | 2/4 | 50 |
| Samples from Different Organs | Standard in 25 mg of Tissues (Copy) | B1 iiPCR No. Positive/ No. Tested | B1 qPCR No. Positive/ No. Tested | RE iiPCR No. Positive/ No. Tested | RE qPCR No. Positive/ No. Tested |
|---|---|---|---|---|---|
| Cerebrum | 3 × 103 | 4/4 | 4/4 | 4/4 | 4/4 |
| 3 × 102 | 4/4 | 4/4 | 1/4 | 4/4 | |
| 3 × 101 | 0/4 | 1/4 | 0/4 | 1/4 | |
| 3 × 100 | 0/4 | 1/4 | 0/4 | 0/4 | |
| Cerebellum | 3 × 103 | 4/4 | 4/4 | 4/4 | 4/4 |
| 3 × 102 | 2/4 | 4/4 | 2/4 | 3/4 | |
| 3 × 101 | 2/4 | 3/4 | 0/4 | 0/4 | |
| 3 × 100 | 0/4 | 0/4 | 0/4 | 0/4 | |
| Muscle | 3 × 103 | 4/4 | 4/4 | 4/4 | 4/4 |
| 3 × 102 | 4/4 | 4/4 | 4/4 | 4/4 | |
| 3 × 101 | 0/4 | 3/4 | 0/4 | 3/4 | |
| 3 × 100 | 0/4 | 0/4 | 0/4 | 0/4 | |
| Myocardium | 3 × 103 | 4/4 | 4/4 | 4/4 | 4/4 |
| 3 × 102 | 2/4 | 3/4 | 2/4 | 4/4 | |
| 3 × 101 | 0/4 | 0/4 | 1/4 | 3/4 | |
| 3 × 100 | 0/4 | 0/4 | 0/4 | 1/4 |
| Samples from Different Organs | Standard in 25 mg of Tissues (Copy) | B1/B2M iiPCR No. Positive/ No. Tested | Rate (%) | B1 qPCR No. Positive/ No. Tested | Rate (%) |
|---|---|---|---|---|---|
| Cerebrum | 3 × 107~3 × 103 | 10/10 | 100 | 10/10 | 100 |
| 750 | 20/20 | 100 | 20/20 | 100 | |
| 375 | 4/8 | 50 | 8/8 | 100 | |
| 187.5 | 5/8 | 62.5 | 8/8 | 100 | |
| Cerebellum | 3 × 107~3 × 103 | 10/10 | 100 | 10/10 | 100 |
| 750 | 20/20 | 100 | 20/20 | 100 | |
| 375 | 2/4 | 50 | 4/4 | 100 | |
| 187.5 | 2/4 | 50 | 4/4 | 100 | |
| Muscle | 3 × 107~3 × 103 | 10/10 | 100 | 10/10 | 100 |
| 750 | 20/20 | 100 | 20/20 | 100 | |
| 375 | 4/4 | 100 | 4/4 | 100 | |
| 187.5 | 3/4 | 75 | 4/4 | 100 | |
| Myocardium | 3 × 107~3 × 103 | 30/30 | 100 | 10/10 | 100 |
| 750 | 12/24 | 50 | 24/24 | 100 |
| Samples from Different Organs | B1/B2M iiPCR | B1 qPCR a | Agreement (κ, CI95%) b | ||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| Overall performance | Positive | 152 | 0 | 152 | 92% (0.84[0.78–0.90]) |
| Negative | 24 | 120 | 144 | ||
| Total | 176 | 120 | 296 | ||
| Cerebrum | Positive | 39 | 0 | 39 | 91% (0.82[0.69–0.94]) |
| Negative | 7 | 30 | 37 | ||
| Total | 46 | 30 | 76 | ||
| Cerebellum | Positive | 34 | 0 | 34 | 94% (0.89[0.77–0.99]) |
| Negative | 4 | 30 | 34 | ||
| Total | 38 | 30 | 68 | ||
| Muscle | Positive | 37 | 0 | 37 | 99% (0.97[0.91–1.00]) |
| Negative | 1 | 30 | 31 | ||
| Total | 38 | 30 | 68 | ||
| Myocardium | Positive | 42 | 0 | 42 | 86% (0.71[0.57–0.86]) |
| Negative | 12 | 30 | 42 | ||
| Total | 54 | 30 | 84 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, M.-J.; Yang, W.-C. A Field-Deployable Insulated Isothermal PCR (iiPCR) for the Global Surveillance of Toxoplasma gondii Infection in Cetaceans. Animals 2022, 12, 506. https://doi.org/10.3390/ani12040506
Hsieh M-J, Yang W-C. A Field-Deployable Insulated Isothermal PCR (iiPCR) for the Global Surveillance of Toxoplasma gondii Infection in Cetaceans. Animals. 2022; 12(4):506. https://doi.org/10.3390/ani12040506
Chicago/Turabian StyleHsieh, Meng-Jung, and Wei-Cheng Yang. 2022. "A Field-Deployable Insulated Isothermal PCR (iiPCR) for the Global Surveillance of Toxoplasma gondii Infection in Cetaceans" Animals 12, no. 4: 506. https://doi.org/10.3390/ani12040506
APA StyleHsieh, M.-J., & Yang, W.-C. (2022). A Field-Deployable Insulated Isothermal PCR (iiPCR) for the Global Surveillance of Toxoplasma gondii Infection in Cetaceans. Animals, 12(4), 506. https://doi.org/10.3390/ani12040506





