Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review
Simple Summary
Abstract
1. Introduction
2. Udder Evaluation Criteria: Past and Present
3. Evaluation Criteria Based on Udder Phenotypic Traits
4. Evaluation Criteria Based on Udder Genomic Traits
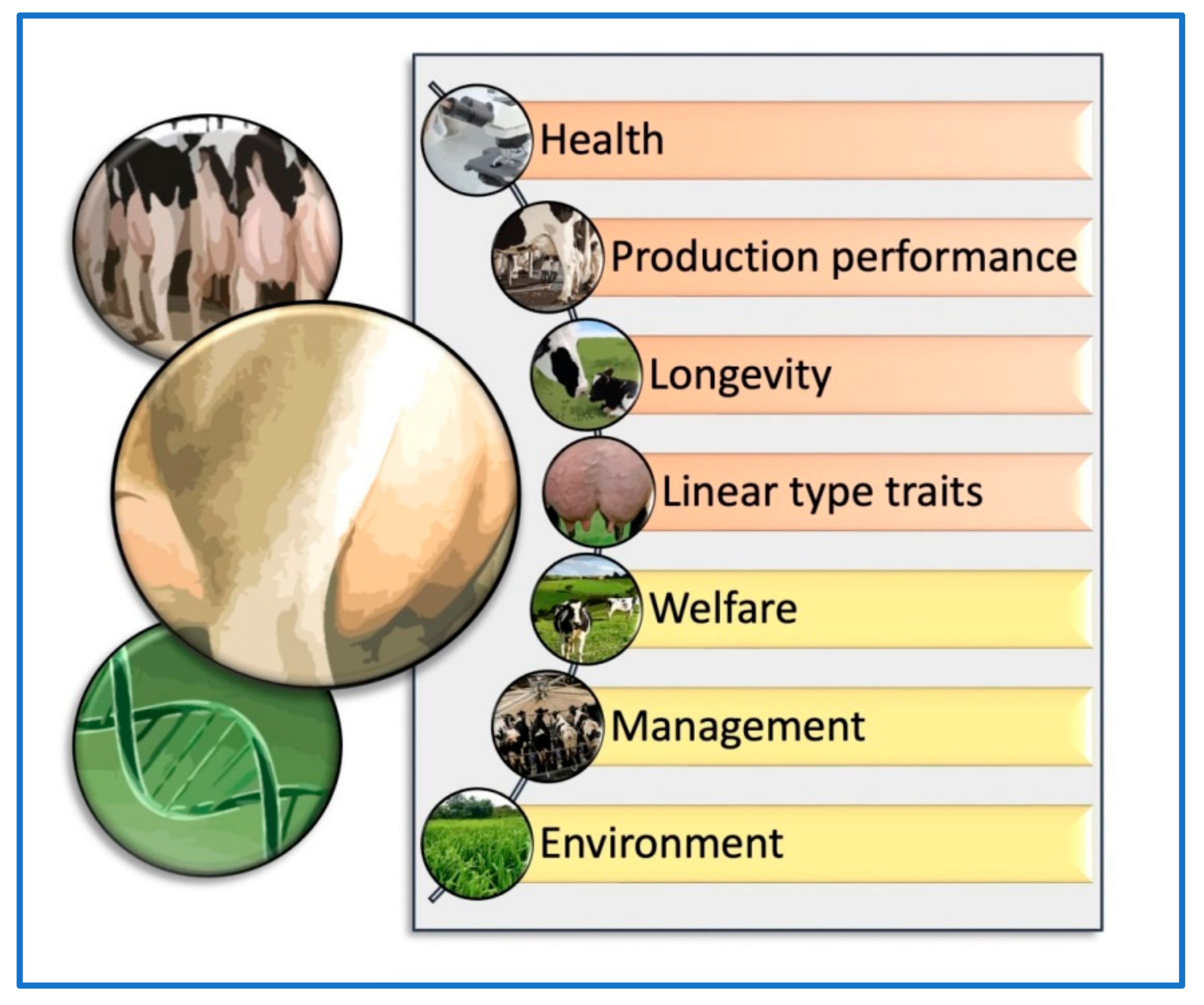
5. Evaluation Criteria Based on Genomic and Phenotypic Traits: Udder Health, Production, and Longevity
5.1. Genomic, Genotypic, and Phenotypic Udder Traits: Impact on Mammary Gland Health
5.2. Genomic, Genotypic, and Phenotypic Udder Traits: Effects on Milk Production and Quality
5.3. Genomic, Genotypic, and Phenotypic Udder Traits: Influence on True and Functional Longevity
5.4. Relationship between Genomic, Genotypic, and Phenotypic Udder Traits: Health, Production, and Longevity
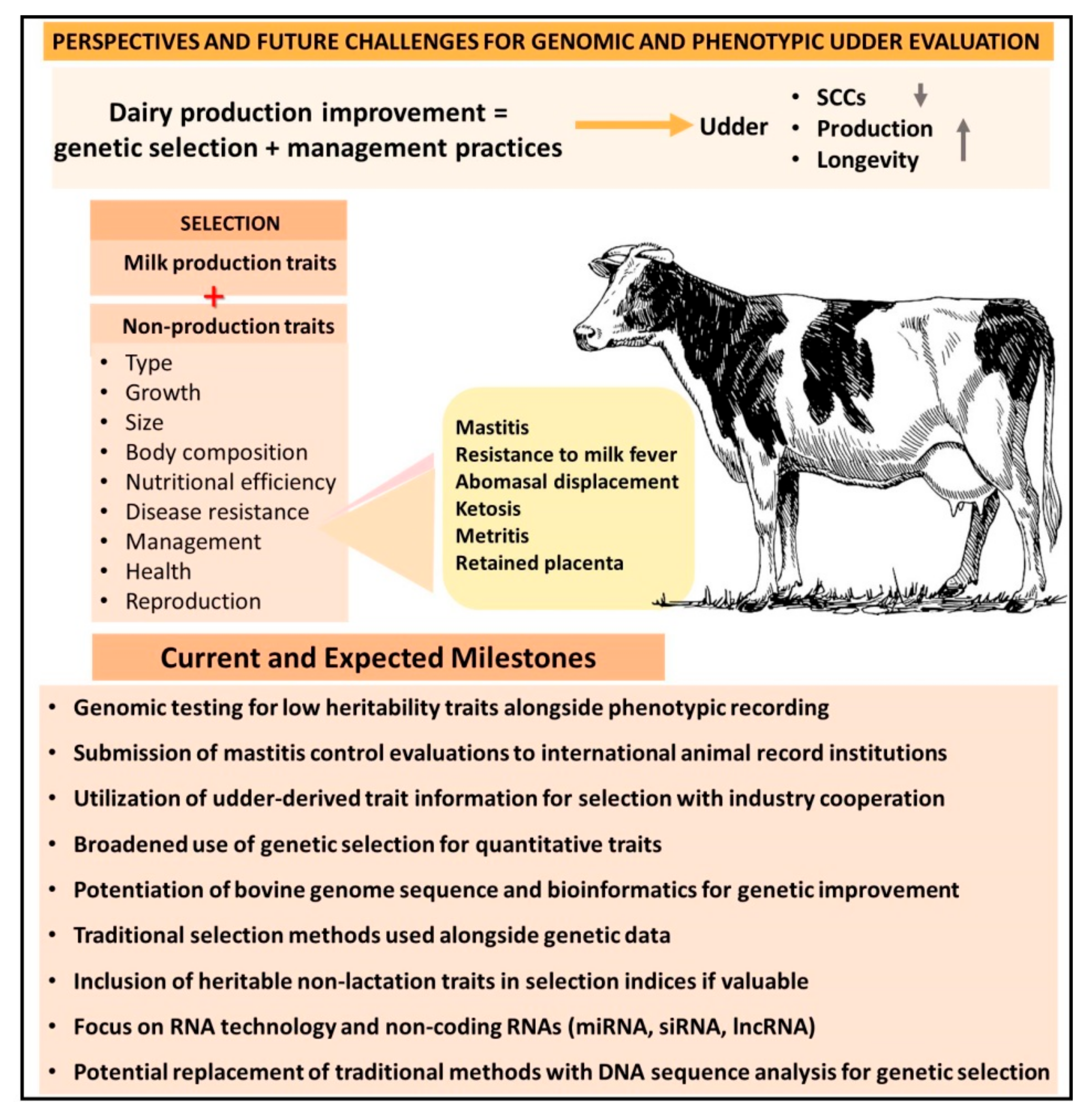
6. Perspectives and Future Challenges for Genomic, Genotypic, and Phenotypic Udder Evaluation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carta, A.; Casu, S.; Salaris, S. Invited Review: Current State of Genetic Improvement in Dairy Sheep. J. Dairy Sci. 2009, 92, 5814–5833. [Google Scholar] [CrossRef] [PubMed]
- Egger-Danner, C.; Cole, J.B.; Pryce, J.E.; Gengler, N.; Heringstad, B.; Bradley, A.; Stock, K.F. Invited Review: Overview of New Traits and Phenotyping Strategies in Dairy Cattle with a Focus on Functional Traits. Animal 2014, 9, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Chesnais, J.P.; Cooper, T.A.; Wiggans, G.R.; Sargolzaei, M.; Pryce, J.E.; Miglior, F. Using Genomics to Enhance Selection of Novel Traits in North American Dairy Cattle. J. Dairy Sci. 2016, 99, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; Cabezas, J.; Rodriguez-Alvarez, L.; Garcia-Herreros, M. Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production-Reproductive Traits. Animals 2020, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Flower, F.C.; Weary, D.M. Gait Assessment in Dairy Cattle. Animal 2009, 3, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.A.; Siegford, J.M. Lactating Dairy Cows Adapt Quickly to Being Milked by an Automatic Milking System. J. Dairy Sci. 2012, 95, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, A.; Macciotta, N.P.P.; Mele, M.; Tagliapietra, F.; Schiavon, S.; Bittante, G.; Pegolo, S. Genetic and Genomic Analyses of Latent Variables Related to the Milk Fatty Acid Profile, Milk Composition, and Udder Health in Dairy Cattle. J. Dairy Sci. 2019, 102, 5254–5265. [Google Scholar] [CrossRef]
- Shanks, R.D.; Rooney, K.A.; Hutjens, M.F. Breeding Practices on Illinois Holstein Farms. J. Dairy Sci. 1983, 66, 1209–1217. [Google Scholar] [CrossRef]
- Beard, J.K.; Musgrave, J.A.; Funston, R.N.; Travis Mulliniks, J. The Effect of Cow Udder Score on Cow/Calf Performance in the Nebraska Sandhills. Transl. Anim. Sci. 2019, 3, 14–19. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. A Review of Inbreeding Depression in Dairy Cattle: Current Status, Emerging Control Strategies, and Future Prospects. J. Dairy Res. 2022, 89, 3–12. [Google Scholar] [CrossRef]
- Mancin, E.; Sartori, C.; Guzzo, N.; Tuliozi, B.; Mantovani, R. Selection Response Due to Different Combination of Antagonistic Milk, Beef, and Morphological Traits in the Alpine Grey Cattle Breed. Animals 2021, 11, 1340. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Bustos, V.J.; Montaldo, H.H.; Valencia-Posadas, M.; Shepard, L.; Pérez-Elizalde, S.; Hernández-Mendo, O.; Torres-Hernández, G. Linear and Nonlinear Genetic Relationships between Type Traits and Productive Life in US Dairy Goats. J. Dairy Sci. 2017, 100, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Hazel, A.R.; Heins, B.J.; Hansen, L.B. Health Treatment Cost, Stillbirth, Survival, and Conformation of Viking Red-, Montbéliarde-, and Holstein-Sired Crossbred Cows Compared with Pure Holstein Cows during Their First 3 Lactations. J. Dairy Sci. 2020, 103, 10917–10939. [Google Scholar] [CrossRef] [PubMed]
- Heimes, A.; Brodhagen, J.; Weikard, R.; Hammon, H.M.; Meyerholz, M.M.; Petzl, W.; Zerbe, H.; Engelmann, S.; Schmicke, M.; Hoedemaker, M.; et al. Characterization of Functional Traits with Focus on Udder Health in Heifers with Divergent Paternally Inherited Haplotypes on BTA18. BMC Vet. Res. 2019, 15, 241. [Google Scholar] [CrossRef]
- Hansen, L.B.; Cole, J.B.; Marx, G.D.; Seykora, A.J. Productive Life and Reasons for Disposal of Holstein Cows Selected for Large versus Small Body Size. J. Dairy Sci. 1999, 82, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Dadati, E.; Kennedy, B.W.; Burnside, E.B. Relationships between Conformation and Calving Interval in Holstein Cows. J. Dairy Sci. 1986, 69, 3112–3119. [Google Scholar] [CrossRef]
- Strandberg, E.; Shook, G.E. Genetic and Economic Responses to Breeding Programs That Consider Mastitis. J. Dairy Sci. 1989, 72, 2136–2142. [Google Scholar] [CrossRef]
- Nazar, M.; Abdalla, I.M.; Chen, Z.; Ullah, N.; Liang, Y.; Chu, S.; Xu, T.; Mao, Y.; Yang, Z.; Lu, X. Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle. Animals 2022, 12, 2542. [Google Scholar] [CrossRef]
- Lucy, M.C. Non-Lactational Traits of Importance in Dairy Cows and Applications for Emerging Biotechnologies. N. Z. Vet. J. 2005, 53, 406–415. [Google Scholar] [CrossRef]
- Pérez-Enciso, M. Genomic Relationships Computed from Either Next-Generation Sequence or Array SNP Data. J. Anim. Breed. Genet. 2014, 131, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Silpa, M.V.; König, S.; Sejian, V.; Malik, P.K.; Nair, M.R.R.; Fonseca, V.F.C.; Maia, A.S.C.; Bhatta, R. Climate-Resilient Dairy Cattle Production: Applications of Genomic Tools and Statistical Models. Front. Vet. Sci. 2021, 8, 625189. [Google Scholar] [CrossRef] [PubMed]
- Parker Gaddis, K.L.; VanRaden, P.M.; Cole, J.B.; Norman, H.D.; Nicolazzi, E.; Dürr, J.W. Symposium Review: Development, Implementation, and Perspectives of Health Evaluations in the United States. J. Dairy Sci. 2020, 103, 5354–5365. [Google Scholar] [CrossRef] [PubMed]
- Hazel, A.R.; Heins, B.J.; Hansen, L.B. Fertility, Survival, and Conformation of Montbéliarde × Holstein and Viking Red × Holstein Crossbred Cows Compared with Pure Holstein Cows during First Lactation in 8 Commercial Dairy Herds. J. Dairy Sci. 2017, 100, 9447–9458. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Hubbard, S.M. Genetic Evaluation of Yield and Type Traits of Dairy Goats in the United States. J. Dairy Sci. 2001, 84, 69–73. [Google Scholar] [CrossRef]
- Casu, S.; Pernazza, I.; Carta, A. Feasibility of a Linear Scoring Method of Udder Morphology for the Selection Scheme of Sardinian Sheep. J. Dairy Sci. 2006, 89, 2200–2209. [Google Scholar] [CrossRef]
- Kelm, S.C.; Freeman, A.E.; Brundage, A.L.; Pearson, R.E.; Martin, T.G.; McGilliard, L.D.; Hansen, L.B.; Young, C.W.; Voelker, H.H.; Shook, G.E.; et al. Direct and Correlated Responses to Selection for Milk Yield: Results and Conclusions of Regional Project NC-2, “Improvement of Dairy Cattle through Breeding, with Emphasis on Selection”. J. Dairy Sci. 2000, 83, 2721–2732. [Google Scholar] [CrossRef]
- Varona, L.; Moreno, C.; Altarriba, J. Genetic Correlation of Longevity with Growth, Post-Mortem, Docility and Some Morphological Traits in the Pirenaica Beef Cattle Breed. Animal 2012, 6, 873–879. [Google Scholar] [CrossRef]
- Persson Waller, K.; Persson, Y.; Nyman, A.K.; Stengärde, L. Udder Health in Beef Cows and Its Association with Calf Growth. Acta Vet. Scand. 2014, 56, 9. [Google Scholar] [CrossRef]
- Just, A.; Wellmann, R.; Bennewitz, J. Estimation of Relative Economic Weights and the Marginal Willingness to Pay for Breeding Traits of Brown Swiss Cattle Using Discrete Choice Experiments. J. Dairy Sci. 2018, 101, 5207–5213. [Google Scholar] [CrossRef]
- Veerkamp, R.F.; Gerritsen, C.L.M.; Koenen, E.P.C.; Hamoen, A.; De Jong, G. Evaluation of Classifiers That Score Linear Type Traits and Body Condition Score Using Common Sires. J. Dairy Sci. 2002, 85, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Buaban, S.; Prempree, S.; Sumreddee, P.; Duangjinda, M.; Masuda, Y. Genomic Prediction of Milk-Production Traits and Somatic Cell Score Using Single-Step Genomic Best Linear Unbiased Predictor with Random Regression Test-Day Model in Thai Dairy Cattle. J. Dairy Sci. 2021, 104, 12713–12723. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, S.; Kumar, S.; Kumar, M. Current Status of Research on Linear Type Traits in Indian Cattle and Future Strategies. Trop. Anim. Health Prod. 2020, 52, 2221–2232. [Google Scholar] [CrossRef] [PubMed]
- Manafiazar, G.; Goonewardene, L.; Miglior, F.; Crews, D.H.; Basarab, J.A.; Okine, E.; Wang, Z. Genetic and Phenotypic Correlations among Feed Efficiency, Production and Selected Conformation Traits in Dairy Cows. Animal 2015, 10, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Miglior, F.; Koeck, A.; Jamrozik, J.; Schenkel, F.; Kelton, D.; Kistemaker, G.; Van Doormaal, B. Index for Mastitis Resistance and Use of BHBA for Evaluation of Health Traits in Canadian Holsteins. Interbull Bull. 2014, 48, 73–78. [Google Scholar]
- Parker Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. Genomic Selection for Producer-Recorded Health Event Data in US Dairy Cattle. J. Dairy Sci. 2014, 97, 3190–3199. [Google Scholar] [CrossRef]
- Rupp, R.; Boichard, D. Genetics of Resistance to Mastitis in Dairy Cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef]
- Sinha, R.; Sinha, B.; Kumari, R.; Vineeth, M.R.; Sharma, N.; Verma, A.; Gupta, I.D. Association of Udder Type Traits with Single Nucleotide Polymorphisms in Sahiwal (Bos indicus) and Karan Fries (Bos taurus × Bos indicus) Cattle. Anim. Biotechnol. 2022, 1–12. [Google Scholar] [CrossRef]
- Pausch, H.; Emmerling, R.; Schwarzenbacher, H.; Fries, R. A Multi-Trait Meta-Analysis with Imputed Sequence Variants Reveals Twelve QTL for Mammary Gland Morphology in Fleckvieh Cattle. Genet. Sel. Evol. 2016, 48, 14. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Zwald, N.R.; Weigel, K.A.; Chang, Y.M.; Welper, R.D.; Clay, J.S. Genetic Selection for Health Traits Using Producer-Recorded Data. II. Genetic Correlations, Disease Probabilities, and Relationships with Existing Traits. J. Dairy Sci. 2004, 87, 4295–4302. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Klemetsdal, G.; Steine, T. Selection Responses for Disease Resistance in Two Selection Experiments with Norwegian Red Cows. J. Dairy Sci. 2007, 90, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Jamrozik, J.; Koeck, A.; Miglior, F.; Kistemaker, G.; Schenkel, F.; Kelton, D.; Doormaal, B. Van Genetic and Genomic Evaluation of Mastitis Resistance in Canada. Interbull Bull. 2013, 47. [Google Scholar]
- Koeck, A.; Miglior, F.; Kelton, D.F.; Schenkel, F.S. Alternative Somatic Cell Count Traits to Improve Mastitis Resistance in Canadian Holsteins. J. Dairy Sci. 2012, 95, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.H. Traits for Sire Selection Related to Udder Health and Management. J. Dairy Sci. 1984, 67, 459–471. [Google Scholar] [CrossRef]
- Thomas, C.L.; Vinson, W.E.; Pearson, R.E.; Dickinson, F.N.; Johnson, L.P. Relationships between Linear Type Scores, Objective Type Measures, and Indicators of Mastitis. J. Dairy Sci. 1984, 67, 1281–1292. [Google Scholar] [CrossRef]
- Ekman, L.; Nyman, A.K.; Landin, H.; Magnusson, U.; Waller, K.P. Mild and Severe Udder Cleft Dermatitis-Prevalence and Risk Factors in Swedish Dairy Herds. J. Dairy Sci. 2018, 101, 556–571. [Google Scholar] [CrossRef]
- Marete, A.; Lund, M.S.; Boichard, D.; Ramayo-Caldas, Y. A System-Based Analysis of the Genetic Determinism of Udder Conformation and Health Phenotypes across Three French Dairy Cattle Breeds. PLoS ONE 2018, 13, e0199931. [Google Scholar] [CrossRef]
- Wagner, P.; Yin, T.; Brügemann, K.; Engel, P.; Weimann, C.; Schlez, K.; König, S. Genome-Wide Associations for Microscopic Differential Somatic Cell Count and Specific Mastitis Pathogens in Holstein Cows in Compost-Bedded Pack and Cubicle Farming Systems. Animals 2021, 11, 1839. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Cui, N.; Liang, Y. Cadherins Associate with Distinct Stem Cell-Related Transcription Factors to Coordinate the Maintenance of Stemness in Triple-Negative Breast Cancer. Stem Cells Int. 2017, 2017, 5091541. [Google Scholar] [CrossRef]
- Li, Y.; Han, B.; Liu, L.; Zhao, F.; Liang, W.; Jiang, J.; Yang, Y.; Ma, Z.; Sun, D. Genetic Association of DDIT3, RPL23A, SESN2 and NR4A1 Genes with Milk Yield and Composition in Dairy Cattle. Anim. Genet. 2019, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Fernando, R.S.; Spahr, S.L.; Jaster, E.H. Comparison of Electrical Conductivity of Milk with Other Indirect Methods for Detection of Subclinical Mastitis. J. Dairy Sci. 1985, 68, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Van Eetvelde, M.; Verdru, K.; de Jong, G.; van Pelt, M.L.; Meesters, M.; Opsomer, G. Researching 100 t Cows: An Innovative Approach to Identify Intrinsic Cows Factors Associated with a High Lifetime Milk Production. Prev. Vet. Med. 2021, 193, 105392. [Google Scholar] [CrossRef] [PubMed]
- Lancelot, R.; Paye, B.; Lescourret, F. Factors Affecting the Distribution of Clinical Mastitis among Udder Quarters in French Dairy Cows. Vet. Res. 1997, 28, 45–53. [Google Scholar] [PubMed]
- Bhutto, A.L.; Murray, R.D.; Woldehiwet, Z. Udder Shape and Teat-End Lesions as Potential Risk Factors for High Somatic Cell Counts and Intra-Mammary Infections in Dairy Cows. Vet. J. 2010, 183, 63–67. [Google Scholar] [CrossRef]
- Guarín, J.F.; Baumberger, C.; Ruegg, P.L. Anatomical Characteristics of Teats and Premilking Bacterial Counts of Teat Skin Swabs of Primiparous Cows Exposed to Different Types of Bedding. J. Dairy Sci. 2017, 100, 1436–1444. [Google Scholar] [CrossRef]
- Miller, R.H.; Pearson, R.E.; Rothschild, M.F.; Fulton, L.A. Comparison of Single and Multiple-Trait Selected Sires. Response in Mastitis Traits. J. Dairy Sci. 1981, 64, 832–837. [Google Scholar] [CrossRef]
- Boettcher, P.J.; Dekkers, J.C.M.; Kolstad, B.W. Development of an Udder Health Index for Sire Selection Based on Somatic Cell Score, Udder Conformation, and Milking Speed. J. Dairy Sci. 1998, 81, 1157–1168. [Google Scholar] [CrossRef]
- Martin, P.; Barkema, H.W.; Brito, L.F.; Narayana, S.G.; Miglior, F. Symposium Review: Novel Strategies to Genetically Improve Mastitis Resistance in Dairy Cattle. J. Dairy Sci. 2018, 101, 2724–2736. [Google Scholar] [CrossRef]
- Van der Geer, D.; Grommers, F.J.; van Houten, M. Comparison of Dairy Cows with Low or High Rate of Udder Infection. Tijdschr. Diergeneeskd. 1979, 104. [Google Scholar] [CrossRef]
- Huntley, S.J.; Cooper, S.; Bradley, A.J.; Green, L.E. A Cohort Study of the Associations between Udder Conformation, Milk Somatic Cell Count, and Lamb Weight in Suckler Ewes. J. Dairy Sci. 2012, 95, 5001–5010. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.; Karrow, N.A.; Thibault, C.; St-Pierre, J.; Lessard, M.; Bissonnette, N. Osteopontin: An Early Innate Immune Marker of Escherichia Coli Mastitis Harbors Genetic Polymorphisms with Possible Links with Resistance to Mastitis. BMC Genom. 2009, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Bharti, P.; Bhakat, C.; Pankaj, P.K.; Bhat, S.A.; Arul Prakash, M.; Thul, M.R.; Puhle Japheth, K. Relationship of Udder and Teat Conformation with Intra-Mammary Infection in Crossbred Cows under Hot-Humid Climate. Vet. World 2015, 8, 898–901. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; Fehr, K.B.; Sepehri, S.; Francoz, D.; De Buck, J.; Barkema, H.W.; Plaizier, J.C.; Khafipour, E. Invited Review: Microbiota of the Bovine Udder: Contributing Factors and Potential Implications for Udder Health and Mastitis Susceptibility. J. Dairy Sci. 2018, 101, 10605–10625. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, A.; Yang, Y.; Yang, C.; Akhtar, M.; Guo, Y.; Shaukat, A.; Guo, M.Y.; Deng, G. Gas6 Negatively Regulates the Staphylococcus Aureus-Induced Inflammatory Response via TLR Signaling in the Mouse Mammary Gland. J. Cell. Physiol. 2020, 235, 7081–7093. [Google Scholar] [CrossRef]
- Van Acker, H.H.; Capsomidis, A.; Smits, E.L.; Van Tendeloo, V.F. CD56 in the Immune System: More Than a Marker for Cytotoxicity? Front. Immunol. 2017, 8, 892. [Google Scholar] [CrossRef]
- Bobbo, T.; Biffani, S.; Taccioli, C.; Penasa, M.; Cassandro, M. Comparison of Machine Learning Methods to Predict Udder Health Status Based on Somatic Cell Counts in Dairy Cows. Sci. Rep. 2021, 11, 13642. [Google Scholar] [CrossRef]
- Bobbo, T.; Matera, R.; Pedota, G.; Manunza, A.; Cotticelli, A.; Neglia, G.; Biffani, S. Exploiting Machine Learning Methods with Monthly Routine Milk Recording Data and Climatic Information to Predict Subclinical Mastitis in Italian Mediterranean Buffaloes. J. Dairy Sci. 2023, 106, 1942–1952. [Google Scholar] [CrossRef]
- Rodenburg, J. Robotic Milking: Technology, Farm Design, and Effects on Work Flow. J. Dairy Sci. 2017, 100, 7729–7738. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive Analysis of Machine Learning Models for Prediction of Sub-Clinical Mastitis: Deep Learning and Gradient-Boosted Trees Outperform Other Models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, C.; Wang, H.; Xu, W.; Zhao, Z.; Chen, M.; Jia, B.; Huang, B. The Early Prediction of Common Disorders in Dairy Cows Monitored by Automatic Systems with Machine Learning Algorithms. Animals 2022, 12, 1251. [Google Scholar] [CrossRef] [PubMed]
- Sawa, A.; Bogucki, M.; Krężel-Czopek, S.; Neja, W. Relationship between Conformation Traits and Lifetime Production Efficiency of Cows. ISRN Vet. Sci. 2013, 2013, 124690. [Google Scholar] [CrossRef] [PubMed]
- Jeretina, J.; Skok, D.J. Genes Associated with Somatic Cell Count Index in Brown Swiss Cattle. J. Anim. Sci. 2020, 98, skaa330. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Koop, G.; Getaneh, A.M.; Lam, T.J.G.M.; Hogeveen, H. Failure Costs Associated with Mastitis in Smallholder Dairy Farms Keeping Holstein Friesian × Zebu Crossbreed Cows. Animal 2019, 13, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Knob, D.A.; Scholz, A.M.; Alessio, D.R.M.; Mendes, B.P.B.; Perazzoli, L.; Kappes, R.; Thaler Neto, A. Reproductive and Productive Performance, Udder Health, and Conformation Traits of Purebred Holstein, F1, and R1 Crossbred Holstein × Simmental Cows. Trop. Anim. Health Prod. 2020, 52, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Gao, J.; Zhao, X.; Ma, Y. Digital Gene Expression Analyses of Mammary Glands from Meat Ewes Naturally Infected with Clinical Mastitis. R. Soc. Open Sci. 2019, 6, 181604. [Google Scholar] [CrossRef]
- Lemay, D.G.; Hovey, R.C.; Hartono, S.R.; Hinde, K.; Smilowitz, J.T.; Ventimiglia, F.; Schmidt, K.A.; Lee, J.W.S.; Islas-Trejo, A.; Silva, P.I.; et al. Sequencing the Transcriptome of Milk Production: Milk Trumps Mammary Tissue. BMC Genom. 2013, 14, 872. [Google Scholar] [CrossRef]
- Raschia, M.A.; Nani, J.P.; Maizon, D.O.; Beribe, M.J.; Amadio, A.F.; Poli, M.A. Single Nucleotide Polymorphisms in Candidate Genes Associated with Milk Yield in Argentinean Holstein and Holstein x Jersey Cows. J. Anim. Sci. Technol. 2018, 60, 31. [Google Scholar] [CrossRef]
- Lashneva, I.; Sermyagin, A.A.; Ignatieva, L.P.; Gladyr, E.; Ermilov, A.; Zinovieva, N.A. PSXII-7 Milk Somatic Cells Monitoring in Russian Holstein Cattle Population as a Base for Determining Genetic and Genomic Variability. J. Anim. Sci. 2021, 99, 252. [Google Scholar] [CrossRef]
- Boettcher, P.J.; Jairath, L.K.; Koots, K.R.; Dekkers, J.C.M. Effects of Interactions between Type and Milk Production on Survival Traits of Canadian Holsteins. J. Dairy Sci. 1997, 80, 2984–2995. [Google Scholar] [CrossRef]
- Zavadilová, L.; Němcová, E.; Štípková, M. Effect of Type Traits on Functional Longevity of Czech Holstein Cows Estimated from a Cox Proportional Hazards Model. J. Dairy Sci. 2011, 94, 4090–4099. [Google Scholar] [CrossRef]
- Sasaki, O. Estimation of Genetic Parameters for Longevity Traits in Dairy Cattle: A Review with Focus on the Characteristics of Analytical Models. Anim. Sci. J. 2013, 84, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Forabosco, F.; Groen, A.F.; Bozzi, R.; Van Arendonk, J.A.M.; Filippini, F.; Boettcher, P.; Bijma, P. Phenotypic Relationships between Longevity, Type Traits, and Production in Chianina Beef Cattle. J. Anim. Sci. 2004, 82, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Hocking, P.M.; McAllister, A.J.; Wolynetz, M.S.; Batra, T.R.; Lee, A.J.; Lin, C.Y.; Roy, G.L.; Vesely, J.A.; Wauthy, J.M.; Winter, K.A. Factors Affecting Length of Herdlife in Purebred and Crossbred Dairy Cattle. J. Dairy Sci. 1988, 71, 1011–1024. [Google Scholar] [CrossRef]
- Ashwell, M.S.; Van Tassell, C.P.; Sonstegard, T.S. A Genome Scan to Identify Quantitative Trait Loci Affecting Economically Important Traits in a US Holstein Population. J. Dairy Sci. 2001, 84, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Szyda, J.; Morek-Kopeć, M.; Komisarek, J.; Zarnecki, A. Evaluating Markers in Selected Genes for Association with Functional Longevity of Dairy Cattle. BMC Genet. 2011, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, T.; Yin, T.; König, S. Survival Analyses in Holstein Cows Considering Direct Disease Diagnoses and Specific SNP Marker Effects. J. Dairy Sci. 2020, 103, 8257–8273. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Cheng, Z.; Cooke, J.S.; Werling, D.; Wathes, D.C.; Pollott, G.E. Polymorphisms in the Selectin Gene Cluster Are Associated with Fertility and Survival Time in a Population of Holstein Friesian Cows. PLoS ONE 2017, 12, e0175555. [Google Scholar] [CrossRef]
- Stefanowska, J.; Plavsic, M.; Ipema, A.H.; Hendriks, M.M.W.B. The Effect of Omitted Milking on the Behaviour of Cows in the Context of Cluster Attachment Failure during Automatic Milking. Appl. Anim. Behav. Sci. 2000, 67, 277–291. [Google Scholar] [CrossRef]
- Kern, E.L.; Cobuci, J.A.; Costa, C.N.; Pimentel, C.M.M.M. Factor Analysis of Linear Type Traits and Their Relation with Longevity in Brazilian Holstein Cattle. Asian-Australas. J. Anim. Sci. 2014, 27, 784–790. [Google Scholar] [CrossRef]
- Short, T.H.; Lawlor, T.J. Genetic Parameters of Conformation Traits, Milk Yield, and Herd Life in Holsteins. J. Dairy Sci. 1992, 75, 1987–1998. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.; Sleator, R.D.; Murphy, C.P.; McCarthy, J.; Berry, D.P. Re-Assessing the Importance of Linear Type Traits in Predicting Genetic Merit for Survival in an Aging Holstein-Friesian Dairy Cow Population. J. Dairy Sci. 2022, 105, 7550–7563. [Google Scholar] [CrossRef] [PubMed]
- Malchiodi, F.; Jamrozik, J.; Christen, A.M.; Fleming, A.; Kistemaker, G.J.; Richardson, C.; Daniel, V.; Kelton, D.F.; Schenkel, F.S.; Miglior, F. Symposium Review: Multiple-Trait Single-Step Genomic Evaluation for Hoof Health. J. Dairy Sci. 2020, 103, 5346–5353. [Google Scholar] [CrossRef]
- Rainard, P.; Foucras, G.; Boichard, D.; Rupp, R. Invited Review: Low Milk Somatic Cell Count and Susceptibility to Mastitis. J. Dairy Sci. 2018, 101, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; VanRaden, P.M. Symposium Review: Possibilities in an Age of Genomics: The Future of Selection Indices. J. Dairy Sci. 2018, 101, 3686–3701. [Google Scholar] [CrossRef]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-Wide Association Analysis of Thirty One Production, Health, Reproduction and Body Conformation Traits in Contemporary U.S. Holstein Cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Sahana, G.; Lund, M.S.; Andersson-Eklund, L.; Hastings, N.; Fernandez, A.; Iso-Touru, T.; Thomsen, B.; Viitala, S.; Sørensen, P.; Williams, J.L.; et al. Fine-Mapping QTL for Mastitis Resistance on BTA9 in Three Nordic Red Cattle Breeds. Anim. Genet. 2008, 39, 354–362. [Google Scholar] [CrossRef]
- Cremonesi, P.; Severgnini, M.; Romanò, A.; Sala, L.; Luini, M.; Castiglioni, B. Bovine Milk Microbiota: Comparison among Three Different DNA Extraction Protocols to Identify a Better Approach for Bacterial Analysis. Microbiol. Spectr. 2021, 9, e00374-21. [Google Scholar] [CrossRef]
- Gryaznova, M.V.; Syromyatnikov, M.Y.; Dvoretskaya, Y.D.; Solodskikh, S.A.; Klimov, N.T.; Mikhalev, V.I.; Zimnikov, V.I.; Mikhaylov, E.V.; Popov, V.N. Microbiota of Cow’s Milk with Udder Pathologies. Microorganisms 2021, 9, 1974. [Google Scholar] [CrossRef]
- Rainard, P. Mammary Microbiota of Dairy Ruminants: Fact or Fiction? Vet. Res. 2017, 48, 25. [Google Scholar] [CrossRef]
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Sultana, M.; Crandall, K.A.; Siddiki, A.Z.; Hossain, M.A. Metagenomic Deep Sequencing Reveals Association of Microbiome Signature with Functional Biases in Bovine Mastitis. Sci. Rep. 2019, 9, 13536. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, R.N.; Middleton, J.R.; McDougall, S.; Katholm, J.; Schukken, Y.H. Molecular Epidemiology of Mastitis Pathogens of Dairy Cattle and Comparative Relevance to Humans. J. Mammary Gland Biol. Neoplasia 2011, 16, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of Seasonal Housing and Teat Preparation on Raw Milk Microbiota: A High-Throughput Sequencing Study. Appl. Environ. Microbiol. 2016, 83, AEM-02694. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.G.; Sanders, J.O.; Knutson, R.E.; Lunt, D.K. Comparison of F1 Bos Indicus x Hereford Cows in Central Texas: II. Udder, Mouth, Longevity, and Lifetime Productivity. J. Anim. Sci. 2001, 79, 1439–1449. [Google Scholar] [CrossRef]
- Tolleson, M.W.; Gill, C.A.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Sanders, J.O.; Riley, D.G. Association of Udder Traits with Single Nucleotide Polymorphisms in Crossbred Bos Indicus- Bos Taurus Cows. J. Anim. Sci. 2017, 95, 2399–2407. [Google Scholar] [CrossRef]
- Mingoas, K.J.P.; Awah-Ndukum, J.; Dakyang, H.; Zoli, P.A. Effects of Body Conformation and Udder Morphology on Milk Yield of Zebu Cows in North Region of Cameroon. Vet. World 2017, 10, 901. [Google Scholar] [CrossRef][Green Version]
- Ventorp, M.; Michanek, P. The Importance of Udder and Teat Conformation for Teat Seeking by the Newborn Calf. J. Dairy Sci. 1992, 75, 262–268. [Google Scholar] [CrossRef]
- Raven, L.A.; Cocks, B.G.; Kemper, K.E.; Chamberlain, A.J.; Vander Jagt, C.J.; Goddard, M.E.; Hayes, B.J. Targeted Imputation of Sequence Variants and Gene Expression Profiling Identifies Twelve Candidate Genes Associated with Lactation Volume, Composition and Calving Interval in Dairy Cattle. Mamm. Genome 2016, 27, 81–97. [Google Scholar] [CrossRef]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-Wide Association Study on Milk Production and Somatic Cell Score for Thai Dairy Cattle Using Weighted Single-Step Approach with Random Regression Test-Day Model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef]
- Dudemaine, P.L.; Thibault, C.; Alain, K.; Bissonnette, N. Genetic Variations in the SPP1 Promoter Affect Gene Expression and the Level of Osteopontin Secretion into Bovine Milk. Anim. Genet. 2014, 45, 629–640. [Google Scholar] [CrossRef]
- Miles, A.M.; Posbergh, C.J.; Huson, H.J. Direct Phenotyping and Principal Component Analysis of Type Traits Implicate Novel QTL in Bovine Mastitis through Genome-Wide Association. Animals 2021, 11, 1147. [Google Scholar] [CrossRef] [PubMed]
- Abbasi-Moshaii, B.; Moradi, M.H.; Yin, T.; Rahimi-Mianji, G.; Nejati-Javaremi, A.; König, S. Genome-Wide Scan for Selective Sweeps Identifies Novel Loci Associated with Resistance to Mastitis in German Holstein Cattle. J. Anim. Breed. Genet. 2023, 140, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Detilleux, J.C. Genetic Factors Affecting Susceptibility to Udder Pathogens. Vet. Microbiol. 2009, 134, 157–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic Selection of High-Yielding Dairy Cattle toward Sustainable Farming Systems in a Rapidly Changing World. Animal 2021, 15, 100292. [Google Scholar] [CrossRef] [PubMed]
- Hagan, B.A.; Moro-Mendez, J.; Cue, R.I. Realized Genetic Selection Differentials in Canadian Ayrshire, Jersey, and Brown Swiss Dairy Cattle Populations. J. Dairy Sci. 2021, 104, 1951–1966. [Google Scholar] [CrossRef]
- González, M.E.; González, V.M.; Montaño, M.F.; Medina, G.E.; Mahadevan, P.; Villa, C.; Villa, R. Genome-Wide Association Analysis of Body Conformation Traits in Mexican Holstein Cattle Using a Mix of Sampled and Imputed SNP Genotypes. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Lund, M.S.; Guldbrandtsen, B.; Buitenhuis, A.J.; Thomsen, B.; Bendixen, C. Detection of Quantitative Trait Loci in Danish Holstein Cattle Affecting Clinical Mastitis, Somatic Cell Score, Udder Conformation Traits, and Assessment of Associated Effects on Milk Yield. J. Dairy Sci. 2008, 91, 4028–4036. [Google Scholar] [CrossRef]
- Wu, X.; Lund, M.S.; Sahana, G.; Guldbrandtsen, B.; Sun, D.; Zhang, Q.; Su, G. Association Analysis for Udder Health Based on SNP-Panel and Sequence Data in Danish Holsteins. Genet. Sel. Evol. 2015, 47, 50. [Google Scholar] [CrossRef]
- Fessenden, B.; Weigel, D.J.; Osterstock, J.; Galligan, D.T.; Di Croce, F. Validation of Genomic Predictions for a Lifetime Merit Selection Index for the US Dairy Industry. J. Dairy Sci. 2020, 103, 10414–10428. [Google Scholar] [CrossRef]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Weighting Sequence Variants Based on Their Annotation Increases the Power of Genome-Wide Association Studies in Dairy Cattle. Genet. Sel. Evol. 2019, 51, 20. [Google Scholar] [CrossRef]
- Mesbah-Uddin, M.; Guldbrandtsen, B.; Capitan, A.; Lund, M.S.; Boichard, D.; Sahana, G. Genome-Wide Association Study with Imputed Whole-Genome Sequence Variants Including Large Deletions for Female Fertility in 3 Nordic Dairy Cattle Breeds. J. Dairy Sci. 2022, 105, 1298–1313. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M.; Tooker, M.E.; O’Connell, J.R.; Cole, J.B.; Bickhart, D.M. Selecting Sequence Variants to Improve Genomic Predictions for Dairy Cattle. Genet. Sel. Evol. 2017, 49, 32. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Schrooten, C.; Bovenhuis, H.; Coppieters, W.; Van Arendonk, J.A.M. Whole Genome Scan to Detect Quantitative Trait Loci for Conformation and Functional Traits in Dairy Cattle. J. Dairy Sci. 2000, 83, 795–806. [Google Scholar] [CrossRef]
- Glantz, M.; Lindmark Måsson, H.; Paulsson, M.; Stålhammar, H. Genomic Selection in Relation to Bovine Milk Composition and Processability. J. Dairy Res. 2012, 79, 53–59. [Google Scholar] [CrossRef]
- Jiménez-Montero, J.A.; González-Recio, O.; Alenda, R. Genotyping Strategies for Genomic Selection in Small Dairy Cattle Populations. Animal 2012, 6, 1216–1224. [Google Scholar] [CrossRef]
- Van der Heide, E.M.M.; Veerkamp, R.F.; van Pelt, M.L.; Kamphuis, C.; Ducro, B.J. Predicting Survival in Dairy Cattle by Combining Genomic Breeding Values and Phenotypic Information. J. Dairy Sci. 2020, 103, 556–571. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s Epigenetic Regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef]
- Saenz-de-Juano, M.D.; Silvestrelli, G.; Bauersachs, S.; Ulbrich, S.E. Determining Extracellular Vesicles Properties and MiRNA Cargo Variability in Bovine Milk from Healthy Cows and Cows Undergoing Subclinical Mastitis. BMC Genom. 2022, 23, 189. [Google Scholar] [CrossRef]
- Cintio, M.; Polacchini, G.; Scarsella, E.; Montanari, T.; Stefanon, B.; Colitti, M. MicroRNA Milk Exosomes: From Cellular Regulator to Genomic Marker. Animals 2020, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Roth, M.J.; Moorehead, R.A. The MiR-200 Family in Normal Mammary Gland Development. BMC Dev. Biol. 2021, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Tabashiri, R.; Sharifi, S.; Pakdel, A.; Bakhtiarizadeh, M.R.; Pakdel, M.H.; Tahmasebi, A.; Hercus, C. Genome-Wide Post-Transcriptional Regulation of Bovine Mammary Gland Response to Streptococcus Uberis. J. Appl. Genet. 2022, 63, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.L.; Yang, J.X.; Li, Z.; Liu, H.Y.; Liu, J.X. Progress on the MiRNA Related with Mammary Gland Development and Lactation. Yi Chuan 2013, 35, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T.; Scheeres, N.; Małopolska, M.M.; Murawski, M.; Agustin, T.D.; Ahmadi, B.; Strzałkowska, N.; Rajtar, P.; Micek, P.; Bartlewski, P.M. Associations between Mammary Gland Echotexture and Milk Composition in Cows. Animals 2020, 10, 2005. [Google Scholar] [CrossRef]
- Zaninelli, M.; Redaelli, V.; Luzi, F.; Bronzo, V.; Mitchell, M.; Dell’Orto, V.; Bontempo, V.; Cattaneo, D.; Savoini, G. First Evaluation of Infrared Thermography as a Tool for the Monitoring of Udder Health Status in Farms of Dairy Cows. Sensors 2018, 18, 862. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals 2023, 13, 1588. https://doi.org/10.3390/ani13101588
Gutiérrez-Reinoso MA, Aponte PM, García-Herreros M. Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals. 2023; 13(10):1588. https://doi.org/10.3390/ani13101588
Chicago/Turabian StyleGutiérrez-Reinoso, Miguel A., Pedro M. Aponte, and Manuel García-Herreros. 2023. "Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review" Animals 13, no. 10: 1588. https://doi.org/10.3390/ani13101588
APA StyleGutiérrez-Reinoso, M. A., Aponte, P. M., & García-Herreros, M. (2023). Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals, 13(10), 1588. https://doi.org/10.3390/ani13101588






