Leveraging Public Data to Predict Global Niches and Distributions of Rhizostome Jellyfishes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Curation
2.2. Statistical Models
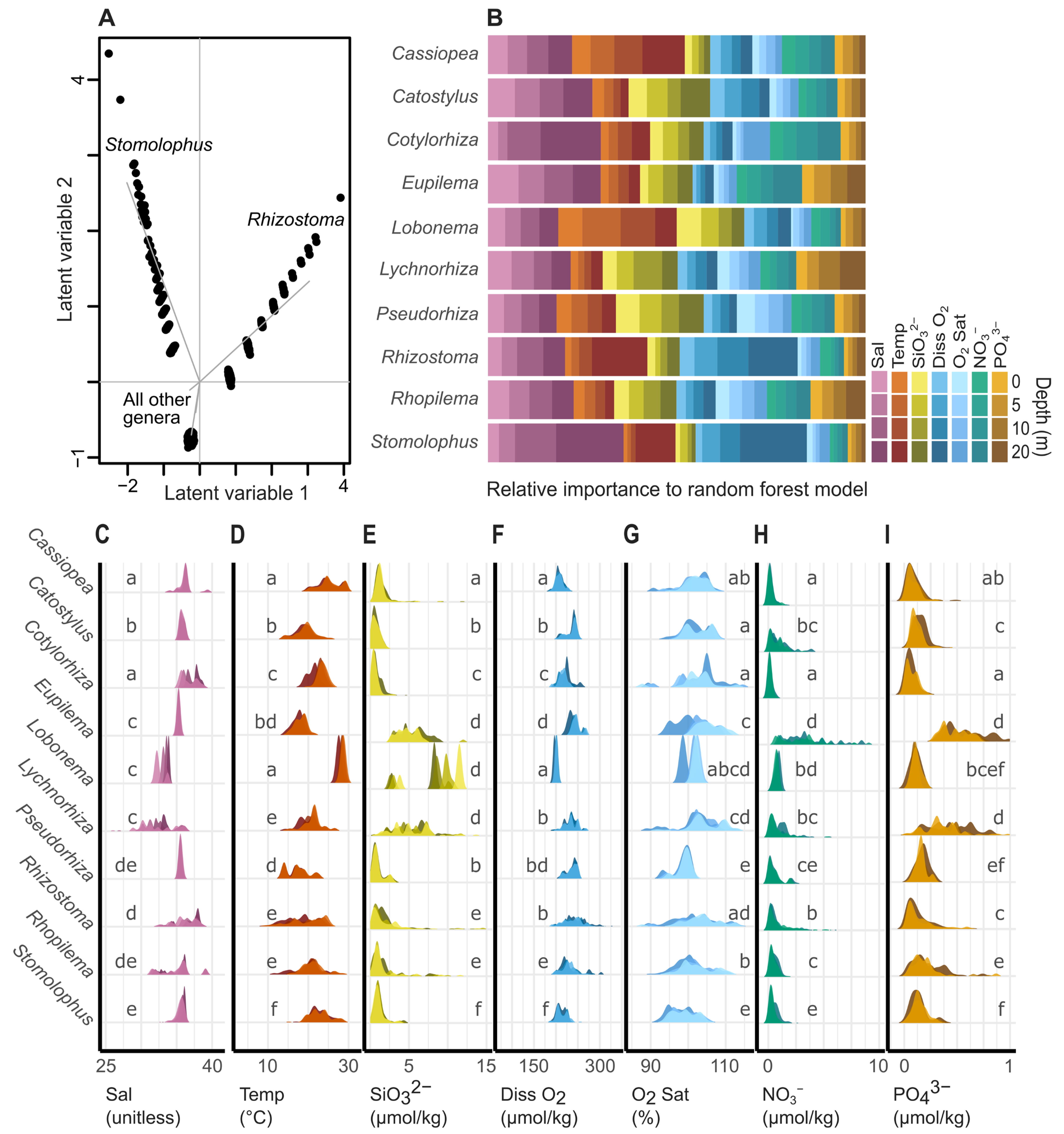
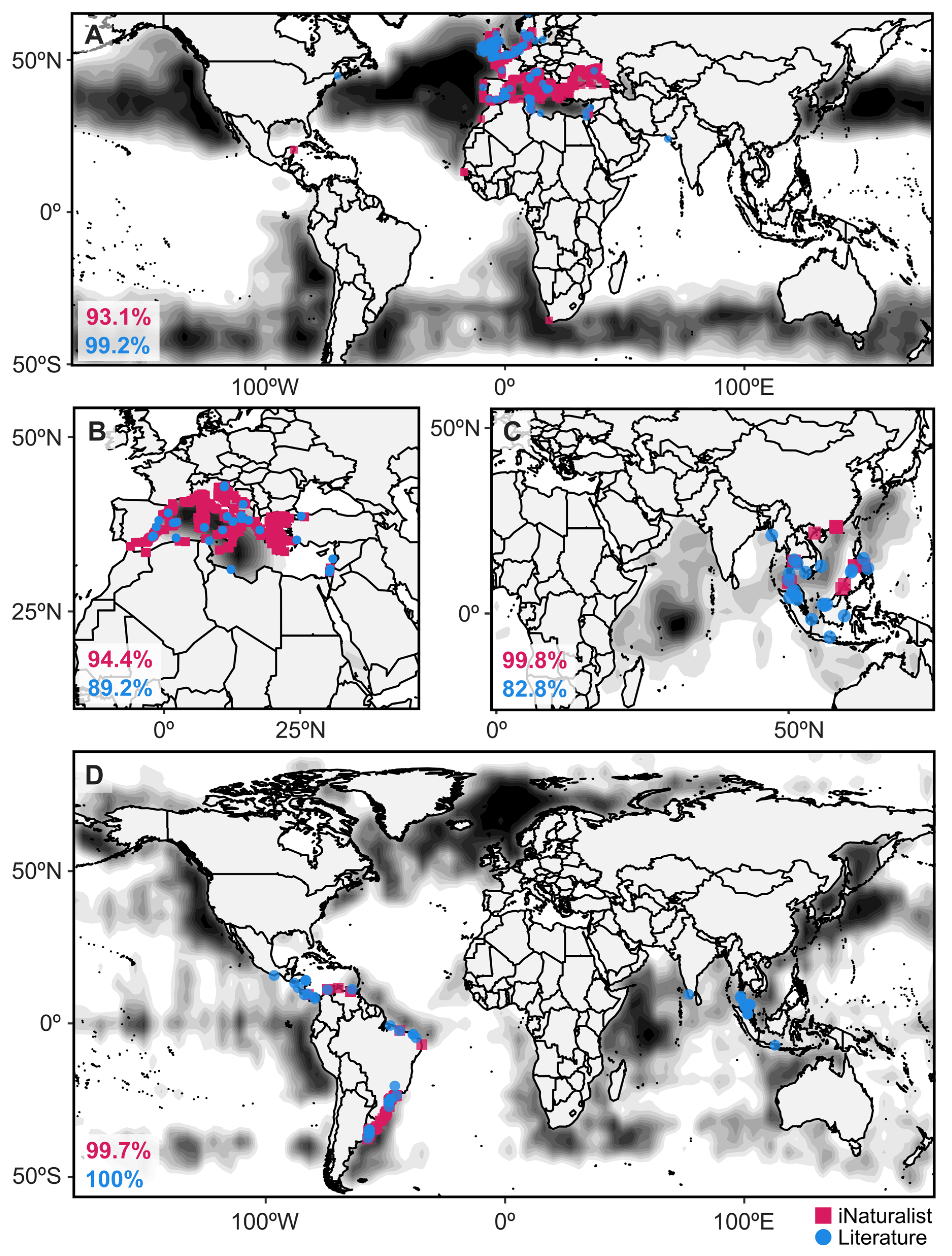
3. Results
3.1. Taxonomic Representation and Distributions of iNaturalist Reports
3.2. Niche Modeling and Distribution Predictions
4. Discussion
4.1. Inferring Ecological Niches
4.2. Predicting Geographic Distributions
4.3. Increasing Global Participation to Reduce Data Biases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelly, R.; Fleming, A.; Pecl, G.; von Gönner, J.; Bonn, A. Citizen science and marine conservation: A global review. Philos. Trans. R. Soc. B 2020, 375, 20190461. [Google Scholar] [CrossRef] [PubMed]
- McKinley, D.C.; Miller-Rushing, A.J.; Ballard, H.L.; Bonney, R.; Brown, H.; Cook-Patton, S.C.; Evans, D.M.; French, R.A.; Parrish, J.K.; Phillips, T.B.; et al. Citizen science can improve conservation science, natural resource management, and environmental protection. Biol. Conserv. 2017, 208, 15–28. [Google Scholar] [CrossRef]
- Cronje, R.; Rohlinger, S.; Crall, A.; Newman, G. Does Participation in Citizen Science Improve Scientific Literacy? A Study to Compare Assessment Methods. Appl. Environ. Educ. Commun. 2011, 10, 135–145. [Google Scholar] [CrossRef]
- Ceccaroni, L.; Bibby, J.; Roger, E.; Flemons, P.; Michael, K.; Fagan, L.; Oliver, J.L. Opportunities and Risks for Citizen Science in the Age of Artificial Intelligence. Citiz. Sci. Theory Pract. 2019, 4, 29. [Google Scholar] [CrossRef]
- Bolt, M.H.; Callaghan, C.T.; Poore, A.G.B.; Verges, A.; Roberts, C.J. Using the background of fish photographs to quantify habitat composition in marine ecosystems. Mar. Ecol. Prog. Ser. 2022, 688, 167–172. [Google Scholar] [CrossRef]
- Soroye, P.; Edwards, B.P.M.; Buxton, R.T.; Ethier, J.P.; Frempong-Manso, A.; Keefe, H.E.; Berberi, A.; Roach-Krajewski, M.; Binley, A.D.; Vincent, J.G.; et al. The risks and rewards of community science for threatened species monitoring. Conserv. Sci. Pract. 2022, 4, e12788. [Google Scholar] [CrossRef]
- Di Cecco, G.J.; Barve, V.; Belitz, M.W.; Stucky, B.J.; Guralnick, R.P.; Hurlbert, A.J. Observing the Observers: How Participants Contribute Data to iNaturalist and Implications for Biodiversity Science. BioScience 2021, 71, 1179–1188. [Google Scholar] [CrossRef]
- Tanaka, K.R.; Van Houtan, K.S.; Mailander, E.; Dias, B.S.; Galginaitis, C.; O’Sullivan, J.; Lowe, C.G.; Jorgensen, S.J. North Pacific warming shifts the juvenile range of a marine apex predator. Sci. Rep. 2021, 11, 3373. [Google Scholar] [CrossRef]
- Helm, R.R. Evolution and development of scyphozoan jellyfish. Biol. Rev. 2018, 93, 1228–1250. [Google Scholar] [CrossRef]
- Graham, W.M.; Pagés, F.; Hamner, W.M. A physical context for gelatinous zooplankton aggregations: A review. Hydrobiologia 2001, 451, 199–212. [Google Scholar] [CrossRef]
- Yasuda, T. Mass occurrence of Stomolophus nomurai was seen again. Umiushi Tsusin 1995, 9, 6–8. (In Japanese) [Google Scholar]
- Lilley, M.K.S.; Houghton, J.D.R.; Hays, G.C. Distribution, extent of inter-annual variability and diet of the bloom-forming jellyfish Rhizostoma in European waters. J. Mar. Biol. Assoc. UK 2009, 89, 39–48. [Google Scholar] [CrossRef]
- Lewis Ames, C. Medusa: A review of an ancient cnidarian body form. In Marine Organisms as Model Systems in Biology and Medicine. Results and Problems in Cell Differentiation; Kloc, M., Kubiak, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; p. 65. [Google Scholar]
- Syazwan, W.M.; Rizman-Idid, M.; Low, L.B.; Then, A.T.-H.; Chong, V.C. Assessment of scyphozoan diversity, distribution and blooms: Implications of jellyfish outbreaks to the environment and human welfare in Malaysia. Reg. Stud. Mar. Sci. 2020, 39, 101444. [Google Scholar] [CrossRef]
- Purcell, J.E. Use of respiration rates of scyphozoan jellyfish to estimate their effects on the food web. Hydrobiologia 2010, 645, 135–152. [Google Scholar] [CrossRef]
- Hays, G.C.; Doyle, T.K.; Houghton, J.D.R. A Paradigm Shift in the Trophic Importance of Jellyfish? Trends Ecol. Evol. 2018, 33, 874–884. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Doyle, T.K.; Creer, S.; Taylor, M.I. Inclusion of jellyfish in 30+ years of Ecopath with Ecosim models. ICES J. Mar. Sci. 2019, 76, 1941–1950. [Google Scholar] [CrossRef]
- Leone, A.; Lecci, R.M.; Miriana, D.; Meli, F.; Piraino, S. The Bright Side of Gelatinous Blooms: Nutraceutical Value and Antioxidant Properties of Three Mediterranean Jellyfish (Scyphozoa). Mar. Drugs 2015, 13, 4654–4681. [Google Scholar] [CrossRef]
- Prieto, L.; Enrique-Navarro, A.; Li Volsi, R.; Ortega, M.J. The large jellyfish Rhizostoma luteum as a sustainable resource for antioxidant properties, nutraceutical value and biomedical applications. Mar. Drugs 2018, 16, 396. [Google Scholar] [CrossRef]
- Muffett, K.M.; Klompen, A.M.L.; Collins, A.G.; Lewis Ames, C. Raising Awareness of the Severity of “Contactless Stings” by Cassiopea Jellyfish and Kin. Animals 2021, 11, 3357. [Google Scholar] [CrossRef]
- Nagata, R.M.; Haddad, M.A.; Nogueira, M., Jr. The nuisance of medusae (Cnidaria, Medusozoa) to shrimp trawls in central part of southern Brazilian Bight, from the perspective of artisanal fishermen. Pan Am. J. Aquat. Sci. 2009, 4, 312–325. [Google Scholar]
- Pauly, D.; Graham, W.; Libralato, S.; Morissette, L.; Palomares, M.L.D. Jellyfish in ecosystems, online databases, and ecosystem models. Hydrobiologia 2009, 61, 67–85. [Google Scholar] [CrossRef]
- Kingsford, M.; Pitt, K.; Gillanders, B. Management of jellyfish fisheries, with special reference to the Order Rhizostomeae. Oceanogr. Mar. Biol. Annu. Rev. 2000, 38, 85–156. [Google Scholar]
- Omori, M.; Nakano, E. Jellyfish fisheries in southeast Asia. Hydrobiologia 2001, 451, 19–26. [Google Scholar] [CrossRef]
- Miyake, H.; Honda, S.; Nishikawa, J.; Yusoff, F.M. Life Cycle of Edible Jellyfish Acromitus hardenbergi Stiasny, 1934 (Scyphozoa: Rhizostomeae) Inhabiting a Brackish-Water Environment. Animals 2021, 11, 2138. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, M.J.; Richardson, A.J. Beyond the jellyfish joyride and global oscillations: Advancing jellyfish research. J. Plankton Res. 2013, 35, 929–938. [Google Scholar] [CrossRef]
- Brotz, L.; Cheung, W.W.L.; Kleisner, K.; Pakhomov, E.; Pauly, D. Increasing jellyfish populations: Trends in Large Marine Ecosystems. Hydrobiologia 2012, 690, 3–20. [Google Scholar] [CrossRef]
- Condon, R.H.; Lucas, C.H.; Pitt, K.A.; Uye, S. Jellyfish blooms and ecological interactions. Mar. Ecol. Prog. Ser. 2014, 510, 109–110. [Google Scholar] [CrossRef]
- Luo, J.Y.; Condon, R.H.; Stock, C.A.; Duarte, C.M.; Lucas, C.H.; Pitt, K.A.; Cowen, R.K. Gelatinous Zooplankton-Mediated Carbon Flows in the Global Oceans: A Data-Driven Modeling Study. Glob. Biogeochem. Cycles 2020, 34, e2020GB006704. [Google Scholar] [CrossRef]
- Cuvier, G. Mémoire sur L’organisation de quelques méduses. Bull. Des Sci. Soc. Philomath. Paris Ser. I Tome 1800, 2, 69. [Google Scholar]
- iNaturalist Observers. iNaturalist Research-Grade Observations. 2023. Available online: https://doi.org/10.15468/ab3s5x (accessed on 3 November 2022).
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Anthony, C.J.; Heagy, M.; Bentlage, B. Phenotypic plasticity in Cassiopea ornata (Cnidaria: Scyphozoa: Rhizostomeae) suggests environmentally driven morphology. Zoomorphology 2022, 141, 115–131. [Google Scholar] [CrossRef]
- Daglio, L.G.; Hayati, R.; Coleman, T.; Han, Y.-M.; Muzaki, F.; Aunurohim; de Bellard, M.E.; Saptarini, D. Species composition of Discomedusae jellyfish (Scyphozoa) in the coastal waters of Eastern Surabaya, East Java. Mar. Biodivers. 2022, 52, 23. [Google Scholar] [CrossRef]
- Lawley, J.W.; Gamero-Mora, E.; Maronna, M.M.; Chiaverano, L.M.; Stampar, S.N.; Hopcroft, R.R.; Collins, A.G.; Morandini, A.C. Morphology is not always useful for diagnosis, and that’s ok: Species hypotheses should not be bound to a class of data. Reply to Brown and Gibbons. S. Afr. J. Sci. 2022, 118, 12590. [Google Scholar] [CrossRef]
- Maronna, M.M.; Lawley, J.W. Current Knowledge on Genetic Diversity of Jellyfishes. In Advances in DNA Barcoding and Molecular Taxonomy-Training manual of International Workshop on DNA Barcoding and Molecular Taxonomy (ITW’04); Nandan, S.B., Lathika, C.T., Aneesh, B., Hari Praved, P., Eds.; Directorate of Public Relations and Publications, CUSAT: Kochi, India, 2022; pp. 36–50. [Google Scholar]
- Locarnini, R.A.; Mishonov, A.V.; Baranova, O.K.; Boyer, T.P.; Zweng, M.M.; Garcia, H.E.; Reagan, J.R.; Seidov, D.; Weathers, K.; Paver, C.R.; et al. Temperature. In World Ocean Atlas 2018; Mishonov, A., Ed.; NOAA Atlas NESDIS 81; National Oceanic and Atmospheric Administration: Silver Spring, MD, USA, 2018; Volume 1, p. 52. [Google Scholar]
- Zweng, M.M.; Reagan, J.R.; Seidov, D.; Boyer, T.P.; Locarnini, R.A.; Garcia, H.E.; Mishonov, A.V.; Baranova, O.K.; Weathers, K.; Paver, C.R.; et al. Salinity. In World Ocean Atlas 2018; Mishonov, A., Ed.; NOAA Atlas NESDIS 82; National Oceanic and Atmospheric Administration: Silver Spring, MD, USA, 2018; Volume 2, p. 50. [Google Scholar]
- Garcia, H.E.; Weathers, K.; Paver, C.R.; Smolyar, I.; Boyer, T.P.; Locarnini, R.A.; Zweng, M.M.; Mishonov, A.V.; Baranova, O.K.; Seidov, D.; et al. Volume 3: Dissolved Oxygen, Apparent Oxygen Utilization, and Oxygen Saturation. In World Ocean Atlas 2018; Mishonov, A., Ed.; NOAA Atlas NESDIS 83; National Oceanic and Atmospheric Administration: Silver Spring, MD, USA, 2018; Volume 3, p. 38. [Google Scholar]
- Garcia, H.E.; Weathers, K.; Paver, C.R.; Smolyar, I.; Boyer, T.P.; Locarnini, R.A.; Zweng, M.M.; Mishonov, A.V.; Baranova, O.K.; Seidov, D.; et al. Dissolved Inorganic Nutrients (phosphate, nitrate and nitrate+nitrite, silicate). In World Ocean Atlas 2018; Mishonov, A., Ed.; NOAA Atlas NESDIS 84; National Oceanic and Atmospheric Administration: Silver Spring, MD, USA, 2018; Volume 4, p. 35. [Google Scholar]
- Boyer, T.P.; Garcia, H.E.; Locarnini, R.A.; Zweng, M.M.; Mishonov, A.V.; Reagan, J.R.; Weathers, K.A.; Baranova, O.K.; Seidov, D.; Smolyar, I.V. World Ocean Atlas 2018 (WOA18); NOAA National Centers for Environmental Information: Boulder, CO, USA, 2018. Available online: https://www.ncei.noaa.gov/archive/accession/NCEI-WOA18 (accessed on 15 December 2022).
- Garcia, H.E.; Boyer, T.P.; Baranova, O.K.; Locarnini, R.A.; Mishonov, A.V.; Grodsky, A.; Paver, C.R.; Weathers, K.W.; Smolyar, L.V.; Reagan, J.R.; et al. World Ocean Atlas 2018: Product Documentation; Mishonov, A., Ed.; NOAA National Centers for Environmental Information: Boulder, CO, USA, 2019.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org (accessed on 3 November 2022).
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2021; Available online: https://rstudio.com (accessed on 3 November 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; Francois, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wang, Y.; Neuman, U.; Wright, S.; Warton, D.I. mvabund: An R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 2012, 3, 471–474. [Google Scholar] [CrossRef]
- Warton, D.I. Many zeros does not mean zero inflation: Comparing the goodness-of-fit of parametric models to multivariate abundance data. Environmetrics 2005, 16, 275–289. [Google Scholar] [CrossRef]
- Warton, D.I. Regularized sandwich estimators for analysis of high dimensional data using generalized estimating equations. Biometrics 2011, 67, 116–123. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, S.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-93. 2022. Available online: https://CRAN.R-project.org/package=caret (accessed on 15 December 2022).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version 1.7-12. 2022. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 15 December 2022).
- Tuszynski, J. caTools: Tools: Moving Window Statistics, GIF, Base64, ROC AUC, etc. R Package Version 1.18.2. 2021. Available online: https://CRAN.R-project.org/package=caTools (accessed on 15 December 2022).
- Valavi, R.; Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J. Predictive performance of presence-only species distribution models: A benchmark study with reproducible code. Ecol. Monogr. 2022, 92, e01486. [Google Scholar] [CrossRef]
- Biau, G. Analysis of Random Forests Model. J. Mach. Learn. Res. 2012, 13, 1063–1095. [Google Scholar]
- Guisan, A.; Edwards, T.C., Jr.; Hastie, T. Generalized linear and generalized additive models in studies of species distributions: Setting the scene. Ecol. Model. 2002, 157, 89–100. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Larcombe, M.J.; Jordan, G.J.; Bryant, D.; Higgins, S.I. The dimensionality of niche space allows bounded and unbounded processes to jointly influence diversification. Nat. Commun. 2018, 9, 4258. [Google Scholar] [CrossRef] [PubMed]
- Quintero, I.; Suchard, M.A.; Jetz, W. Macroevolutionary dynamics of climatic niche space. Proc. R. Soc. B 2022, 289, 20220091. [Google Scholar] [CrossRef]
- Polechová, J.; Storch, D. Ecological niche. In Encyclopedia of Ecology, 2nd ed.; Fath, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 72–80. [Google Scholar] [CrossRef]
- Nagata, R.M.; Teixeira-Amaral, P.; Lemos, V.R.; Jordano, M.A.; Muxagata, E.; Morandini, A.C. First description of wild-collected ephyrae of Lychnorhiza lucerne (Cnidaria, Scyphozoa). An. Acad. Bras. Cienc. 2021, 93, e20190574. [Google Scholar] [CrossRef]
- Nagata, R.M.; Morandini, A.C. Diet, prey selection, and individual feeding rates of the jellyfish Lychnorhiza lucerna (Scyphozoa, Rhizostomeae). Mar. Biol. 2018, 165, 187. [Google Scholar] [CrossRef]
- Bentlage, B.; Peterson, A.T.; Barve, N.; Cartwright, P. Plumbing the depths: Extending ecological niche modelling and species distribution modelling in three dimensions. Glob. Ecol. Biogeogr. 2013, 22, 952–961. [Google Scholar] [CrossRef]
- Duffy, G.; Chown, S. Explicitly integrating a third dimension in marine species distribution modelling. Mar. Ecol. Prog. Ser. 2017, 564, 1–8. [Google Scholar] [CrossRef]
- Owens, H.L.; Rahbek, C. voluModel: Modelling species distributions in three-dimensional space. Methods Ecol. Evol. 2023, 14, 841–847. [Google Scholar] [CrossRef]
- López-Martínez, J.; Arzola-Sotelo, E.A.; Nevárez-Martínez, M.O.; Álvarez-Tello, F.J.; Morales-Bojórquez, E. Modeling growth on the cannonball jellyfish Stomolophus meleagris based on a multi-model inference approach. Hydrobiologia 2020, 847, 1399–1422. [Google Scholar] [CrossRef]
- Giovos, I.; Kleitou, P.; Poursanidis, D.; Batjaka, I.; Bernardi, G.; Crocetta, F.; Doumpas, N.; Kalogirou, S.; Kampouris, T.E.; Keramidas, I.; et al. Citizen-science for monitoring marine invasions and stimulating public engagement: A case project from the eastern Mediterranean. Biol. Invasions 2019, 21, 3707–3721. [Google Scholar] [CrossRef]
- Cordier, J.M.; Loyola, R.; Rojas-Soto, O.; Nori, J. Modeling invasive species risk from established populations: Insights for management and conservation. Perspect. Ecol. Conserv. 2020, 18, 132–138. [Google Scholar] [CrossRef]
- Nogueira, M., Jr.; Haddad, M.A. Macromedusae (Cnidaria) From the Parana Coast, Southern Brazil. J. Coast. Res. 2006, 39, 1161–1164. [Google Scholar]
- Cornelius; Silveira, F.L. Recent Observations on the Brazilian Scyphomedusa Fauna. Resumos VII Congr. Latinoam. Cienc. 1997, 1, 192–193. [Google Scholar]
- Segura-Puertas, L.; Suárez-Morales, E.; Celis, L. A checklist of the Medusae (Hydrozoa, Scyphozoa and Cubozoa) of Mexico. Zootaxa 2003, 194, 1–15. [Google Scholar] [CrossRef]
- Lindsay, D.J.; Grossmann, M.M.; Bentlage, B.; Collins, A.G.; Minemizu, R.; Hopcroft, R.R.; Miyake, H.; Hidaka-Umetsu, M.; Nishikawa, J. The perils of online biogeographic databases: A case study with the ‘monospecific’ genus Aegina (Cnidaria, Hydrozoa, Narcomedusae). Mar. Biol. Res. 2017, 13, 494–512. [Google Scholar] [CrossRef]
- Nogueira, T.A.C.; Ayala, W.E.; Dayrell, J.S.; de Fraga, R.; Kaefer, I.L. Scale-dependent estimates of niche overlap and environmental effects on two sister species of Neotropical snakes. Stud. Neotrop. Fauna Environ. 2019, 54, 121–132. [Google Scholar] [CrossRef]
- Melo-Merino, S.M.; Reyes-Bonilla, H.; Lira-Noriega, A. Ecological niche models and species distribution models in marine environments: A literature review and spatial analysis of evidence. Ecol. Model. 2020, 415, 108837. [Google Scholar] [CrossRef]
- Chauvier, Y.; Descombes, P.; Guéguen, M.; Boulangeat, L.; Thuiller, W.; Zimmermann, N.E. Resolution in species distribution models shapes spatial patterns of plant multifaceted diversity. Ecography 2022, 2022, e05974. [Google Scholar] [CrossRef]
- Gómez-Daglio, L.; Dawson, M.N. Species richness of jellyfishes (Scyphozoa: Discomedusae) in the Tropical Eastern Pacific: Missed taxa, molecules, and morphology match in a biodiversity hotspot. Invertebr. Syst. 2017, 31, 635–663. [Google Scholar] [CrossRef]
- Nature Index. Available online: https://www.nature.com/nature-index/annual-tables/2022/country/all/all (accessed on 15 February 2022).
- Matume, G. Africa aims for a scientific revolution. Afr. Renew. 2007, 21, 10–13. [Google Scholar] [CrossRef]
- Ngongalah, L.; Emerson, W.; Rawlings, N.N.; Musisi, J.M. Research challenges in Africa—An exploratory study on the experiences and opinions of African researchers. bioRxiv 2018. [Google Scholar] [CrossRef]
- Ogunji, J.O.; Prasadam-Halls, B.; Buckley, V. Strengthening research in Africa and improving the competences of African researchers: Lessons from Association of Commonwealth Universities (ACU) projects in Sub-Saharan Africa. In Proceedings of the 7th African Regional Conference of Vice Chancellors and Deans of Science, Education, Engineering and Technology, Victoria Falls, Zimbabwe, 20–21 November 2019. [Google Scholar]
- Kavulya, J.M.; Kyengo, B.N. Research excellence and sustainable development in Africa: An overview of current concerns and prospects in the 21st century. In Proceedings of the 8th Chuka University Interdisciplinary Conference on “Higher Education Strategies for Mitigating Impact of Pandemics (HESMIP)”, Chuka, Kenya, 7–8 October 2021. [Google Scholar]
- Parreira, M.R.; Machado, K.B.; Logares, R.; Diniz-Filho, J.A.F.; Nabout, J.C. The roles of geographic distance and socioeconomic factors on international collaboration among ecologists. Scientometrics 2017, 113, 1539–1550. [Google Scholar] [CrossRef]
- Callaghan, C.T.; Rowley, J.J.L.; Cornwell, W.K.; Poore, A.G.B.; Major, R.E. Improving big citizen science data: Moving beyond haphazard sampling. PLoS Biol. 2019, 17, e3000357. [Google Scholar] [CrossRef] [PubMed]
- Barber, P.H.; Hayes, T.B.; Johnson, T.L.; Márquez-Magaña, L.M. Systemic racism in higher education. Science 2020, 6510, 1440–1441. [Google Scholar] [CrossRef]
- Eisen, M.B. Racism in Science: We need to act now. Elife 2020, 9, e59636. [Google Scholar] [CrossRef] [PubMed]
- Riggs, B. Training matters! Narrative from a Black scientist. MBoC 2021, 32, 223–225. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anthony, C.J.; Tan, K.C.; Pitt, K.A.; Bentlage, B.; Ames, C.L. Leveraging Public Data to Predict Global Niches and Distributions of Rhizostome Jellyfishes. Animals 2023, 13, 1591. https://doi.org/10.3390/ani13101591
Anthony CJ, Tan KC, Pitt KA, Bentlage B, Ames CL. Leveraging Public Data to Predict Global Niches and Distributions of Rhizostome Jellyfishes. Animals. 2023; 13(10):1591. https://doi.org/10.3390/ani13101591
Chicago/Turabian StyleAnthony, Colin Jeffrey, Kei Chloe Tan, Kylie Anne Pitt, Bastian Bentlage, and Cheryl Lewis Ames. 2023. "Leveraging Public Data to Predict Global Niches and Distributions of Rhizostome Jellyfishes" Animals 13, no. 10: 1591. https://doi.org/10.3390/ani13101591




