Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Greek Black Pig Samples
2.2. DNA Extraction and Genotyping of Polymorphisms in the Greek Black Pigs
2.3. Genotyping Data from Other Pig Populations
2.4. Data Analyses
3. Results
3.1. Genotyping Results of Greek Black Pigs
3.1.1. Polymorphisms in the MC1R Gene
3.1.2. Polymorphisms in the Other Genes
3.2. Comparative Analyses with Other Pig Breeds and Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Porter, V. Pigs: A Handbook to the Breeds of the World; Cornell University Press: Ithaca, NY, USA, 1993. [Google Scholar]
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef]
- Larson, G.; Albarella, U.; Dobney, K.; Rowley-Conwy, P.; Schibler, J.; Tresset, A.; Vigne, J.D.; Edwards, C.J.; Schlumbaum, A.; Dinu, A.; et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc. Natl. Acad. Sci. USA 2007, 104, 15276–15281. [Google Scholar] [CrossRef] [PubMed]
- Larson, G.; Liu, R.; Zhao, X.; Yuan, J.; Fuller, D.; Barton, L.; Dobney, K.; Fan, Q.; Gu, Z.; Liu, X.H.; et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc. Natl. Acad. Sci. USA 2010, 107, 7686–7691. [Google Scholar] [CrossRef] [PubMed]
- Čandek-Potokar, M.; Fontanesi, L.; Lebret, B.; Gil, J.M.; Ovilo, C.; Nieto, R.; Fernandez, A.; Pugliese, C.; Oliver, M.; Bozzi, R. Introductory Chapter: Concept and Ambition of Project TREASURE. In European Local Pig Breeds-Diversity and Performance. A Study of Project Treasure; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Monteiro, A.N.T.R.; Wilfart, A.; Utzeri, V.J.; Lukač, N.B.; Tomažin, U.; Nanni Costa, L.; Čandek-Potokar, M.; Fontanesi, L.; Garcia-Launay, F. Environmental impacts of pig production systems using European local breeds: The contribution of carbon sequestration and emissions from grazing. J. Clean. Prod. 2019, 237, 117843. [Google Scholar] [CrossRef]
- Pugliese, C.; Sirtori, F. Quality of meat and meat products produced from southern European pig breeds. Meat Sci. 2012, 90, 511–518. [Google Scholar] [CrossRef]
- Bonneau, M.; Lebret, B. Production systems and influence on eating quality of pork. Meat Sci. 2010, 84, 293–300. [Google Scholar] [CrossRef]
- Muñoz, M.; Bozzi, R.; García, F.; Núñez, Y.; Geraci, C.; Crovetti, A.; García-Casco, J.; Alves, E.; Škrlep, M.; Charneca, R.; et al. Diversity across major and candidate genes in European local pig breeds. PLoS ONE 2018, 13, e0207475. [Google Scholar] [CrossRef]
- Muñoz, M.; Bozzi, R.; García-Casco, J.; Núñez, Y.; Ribani, A.; Franci, O.; García, F.; Škrlep, M.; Schiavo, G.; Bovo, S.; et al. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip. Sci. Rep. 2019, 9, 13546. [Google Scholar] [CrossRef]
- Bovo, S.; Ribani, A.; Muñoz, M.; Alves, E.; Araujo, J.P.; Bozzi, R.; Čandek-Potokar, M.; Charneca, R.; Di Palma, F.; Etherington, G.; et al. Whole-genome sequencing of European autochthonous and commercial pig breeds allows the detection of signatures of selection for adaptation of genetic resources to different breeding and production systems. Genet. Sel. Evol. 2020, 52, 33. [Google Scholar] [CrossRef]
- Bovo, S.; Ribani, A.; Muñoz, M.; Alves, E.; Araujo, J.P.; Bozzi, R.; Charneca, R.; Di Palma, F.; Etherington, G.; Fernandez, A.I.; et al. Genome-wide detection of copy number variants in European autochthonous and commercial pig breeds by whole-genome sequencing of DNA pools identified breed-characterising copy number states. Anim. Genet. 2020, 51, 541–556. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Geraci, C.; Tinarelli, S.; Djan, M.; Veličković, N.; Doneva, R.; Dall’Olio, S.; Nanni Costa, L.; Schiavo, G.; et al. Signatures of de-domestication in autochthonous pig breeds and of domestication in wild boar populations from MC1R and NR6A1 allele distribution. Anim. Genet. 2019, 50, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Antikas, T.G. Pigs in Greece: From Boar (ing) Myths to Pig Epitaphs. In Proceedings of the International Conference―Pigs and Humans, Durham, UK, 26–29 September 2003. [Google Scholar]
- Papatsiros, V.G.; Tassis, P.D.; Christodoulopoulos, G.; Boutsini, S.; Tsirigotakis, G.; Tzika, E.D. Health and production of Greek organic pig farming: Current situation and perspectives. J. Hell. Vet. Med. Soc. 2012, 63, 37–44. [Google Scholar] [CrossRef]
- Michailidou, S.; Kalivas, A.; Ganopoulos, I.; Stea, E.; Michailidis, G.; Tsaftaris, A.; Argiriou, A. A multi-farm assessment of Greek black pig genetic diversity using microsatellite molecular markers. Genet. Mol. Res. 2014, 13, 2752–2765. [Google Scholar] [CrossRef] [PubMed]
- Laliotis, G.P.; Marantidis, A.; Avdi, M. Association of BF, RBP4, and ESR2 genotypes with litter size in an autochthonous pig population. Anim. Biotechnol. 2017, 28, 138–143. [Google Scholar] [CrossRef]
- Laliotis, G.P.; Avdi, M. Evidence of genetic hybridization of the wild boar and the indigenous black pig in Northern Greece. Biotechnol. Anim. Husb. 2018, 34, 149–158. [Google Scholar] [CrossRef]
- Kijas, J.M.; Wales, R.; Törnsten, A.; Chardon, P.; Moller, M.; Andersson, L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998, 150, 1177–1185. [Google Scholar] [CrossRef]
- Kijas, J.M.; Moller, M.; Plastow, G.; Andersson, L.A. A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics 2001, 158, 779–785. [Google Scholar] [CrossRef]
- Fang, M.; Larson, G.; Ribeiro, H.S.; Li, N.; Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 2009, 5, e1000341. [Google Scholar] [CrossRef]
- Fontanesi, L. Genetics and genomics of pigmentation variability in pigs: A review. Livest. Sci. 2022, 265, 105079. [Google Scholar] [CrossRef]
- Johansson Moller, M.; Chaudhary, R.; Hellmén, E.; Höyheim, B.; Chowdhary, B.; Andersson, L. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 1996, 7, 822–830. [Google Scholar] [CrossRef]
- Marklund, S.; Kijas, J.; Rodriguez-Martinez, H.; Rönnstrand, L.; Funa, K.; Moller, M.; Lange, D.; Edfors-Lilja, I.; Andersson, L. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998, 8, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Pielberg, G.; Olsson, C.; Syvänen, A.C.; Andersson, L. Unexpectedly high allelic diversity at the KIT locus causing dominant white color in the domestic pig. Genetics 2002, 160, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; D’Alessandro, E.; Scotti, E.; Liotta, L.; Crovetti, A.; Chiofalo, V.; Russo, V. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim. Genet. 2010, 41, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Scotti, E.; Gallo, M.; Nanni Costa, L.; Dall’Olio, S. Authentication of “mono-breed” pork products: Identification of a coat colour gene marker in Cinta Senese pigs useful to this purpose. Livest. Sci. 2016, 184, 71–77. [Google Scholar] [CrossRef]
- Mikawa, S.; Morozumi, T.; Shimanuki, S.; Hayashi, T.; Uenishi, H.; Domukai, M.; Okumura, N.; Awata, T. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 2007, 17, 586–593. [Google Scholar] [CrossRef]
- Mikawa, S.; Sato, S.; Nii, M.; Morozumi, T.; Yoshioka, G.; Imaeda, N.; Yamaguchi, T.; Hayashi, T.; Awata, T. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 2011, 12, 5. [Google Scholar] [CrossRef]
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Jungerius, B.J.; Van Laere, A.S.; Te Pas, M.F.W.; Van Oost, B.A.; Andersson, L.; Groenen, M.A.M. The IGF2-intron3-G3072A substitution explains a major imprinted QTL effect on backfat thickness in a Meishan x European white pig intercross. Genet. Res. 2004, 84, 95–101. [Google Scholar] [CrossRef]
- Fontanesi, L.; Schiavo, G.; Scotti, E.; Galimberti, G.; Calò, D.G.; Samorè, A.B.; Gallo, M.; Russo, V.; Buttazzoni, L. A retrospective analysis of allele frequency changes of major genes during 20 years of selection in the Italian Large White pig breed. J. Anim. Breed. Genet. 2015, 132, 239–246. [Google Scholar] [CrossRef]
- Tinarelli, S.; Ribani, A.; Utzeri, V.J.; Taurisano, V.; Bovo, C.; Dall’Olio, S.; Nen, F.; Bovo, S.; Schiavo, G.; Gallo, M.; et al. Redefinition of the Mora Romagnola pig breed herd book standard based on DNA markers useful to authenticate its “mono-breed” products: An example of sustainable conservation of a livestock genetic resource. Animals 2021, 11, 526. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory, Cold Spring Harbor: New York, NY, USA, 1989. [Google Scholar]
- Giuffra, E.; Törnsten, A.; Marklund, S.; Bongcam-Rudloff, E.; Chardon, P.; Kijas, J.M.; Anderson, S.I.; Archibald, A.L.; Andersson, L. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome 2002, 13, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Ribani, A.; Scotti, E.; Utzeri, V.J.; Veličković, N.; Dall’Olio, S. Differentiation of meat from European wild boars and domestic pigs using polymorphisms in the MC1R and NR6A1 genes. Meat Sci. 2014, 98, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Linderholm, A.; Spencer, D.; Battista, V.; Frantz, L.; Barnett, R.; Fleischer, R.C.; James, H.F.; Duffy, D.; Sparks, J.P.; Clements, D.R.; et al. A novel MC1R allele for black coat colour reveals the Polynesian ancestry and hybridization patterns of Hawaiian feral pigs. R. Soc. Open Sci. 2016, 3, 160304. [Google Scholar] [CrossRef]
- Fernández, A.; Fabuel, E.; Alves, E.; Rodriguez, C.; Silío, L.; Óvilo, C. DNA tests based on coat colour genes for authentication of the raw material of meat products from Iberian pigs. J. Sci. Food Agric. 2004, 84, 1855–1860. [Google Scholar] [CrossRef]
- Babicz, M.; Pastwa, M.; Skrzypczak, E.; Buczyński, J.T. Variability in the melanocortin 1 receptor (MC1R) gene in wild boars and local pig breeds in Poland. Anim. Genet. 2013, 44, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.C.; Ren, J.; Guo, Y.M.; Ding, N.S.; Chen, C.Y.; Huang, L.S. Genetic evidence for the origin of an IGF2 quantitative trait nucleotide in Chinese pigs. Anim. Genet. 2006, 37, 179–180. [Google Scholar] [CrossRef]
- Ojeda, A.; Huang, L.S.; Ren, J.; Angiolillo, A.; Cho, I.C.; Soto, H.; Lemús-Flores, C.; Makuza, S.M.; Folch, J.M.; Pérez-Enciso, M. Selection in the making: A worldwide survey of haplotypic diversity around a causative mutation in porcine IGF2. Genetics 2008, 178, 1639–1652. [Google Scholar] [CrossRef]
- Domestic Animal Diversity Information System—DAD-IS. 2023. Available online: https://www.fao.org/dad-is/en/ (accessed on 30 March 2023).
- Koutsogiannouli, E.A.; Moutou, K.A.; Sarafidou, T.; Stamatis, C.; Mamuris, Z. Detection of hybrids between wild boars (Sus scrofa scrofa) and domestic pigs (Sus scrofa f. domestica) in Greece, using the PCR-RFLP method on melanocortin-1 receptor (MC1R) mutations. Mamm. Biol. 2010, 75, 69–73. [Google Scholar] [CrossRef]
- Gamborg, C.; Gremmen, B.; Christiansen, S.B.; Sandoe, P. De-domestication: Ethics at the intersection of landscape restoration and animal welfare. Environ. Values 2010, 19, 57–78. [Google Scholar] [CrossRef]
- Thulin, C.G.; Alves, P.C.; Djan, M.; Fontanesi, L.; Peacock, D. Wild opportunities with dedomestication genetics of rabbits. Restor. Ecol. 2017, 25, 330–332. [Google Scholar] [CrossRef]
- Petrelli, S.; Buglione, M.; Maselli, V.; Troiano, C.; Larson, G.; Frantz, L.; Manin, A.; Ricca, E.; Baccigalupi, L.; Wright, D.; et al. Population genomic, olfactory, dietary, and gut microbiota analyses demonstrate the unique evolutionary trajectory of feral pigs. Mol. Ecol. 2022, 31, 220–237. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.; Fontanesi, L.; Davoli, R.; Chiofalo, L.; Liotta, L.; Zumbo, A. Analysis of single nucleotide polymorphisms in major and candidate genes for production traits in Nero Siciliano pig breed. Ital. J. Anim. Sci. 2004, 3, 19–29. [Google Scholar] [CrossRef]
- Rodríguez, C.I.; Setaluri, V. Cyclic AMP (cAMP) signaling in melanocytes and melanoma. Arch. Biochem. Biophys. 2014, 563, 22–27. [Google Scholar] [CrossRef]
- Fan, Y.; Xing, Y.; Zhang, Z.; Ai, H.; Ouyang, Z.; Ouyang, J.; Yang, M.; Li, P.; Chen, Y.; Gao, J.; et al. A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs. PLoS ONE 2013, 8, e62534. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, S.; Ribani, A.; Moscatelli, G.; Zambonelli, P.; Gallo, M.; Nanni Costa, L.; Fontanesi, L. Teat number parameters in Italian Large White pigs: Phenotypic analysis and association with vertnin (VRTN) gene allele variants. Livest. Sci. 2018, 210, 68–72. [Google Scholar] [CrossRef]

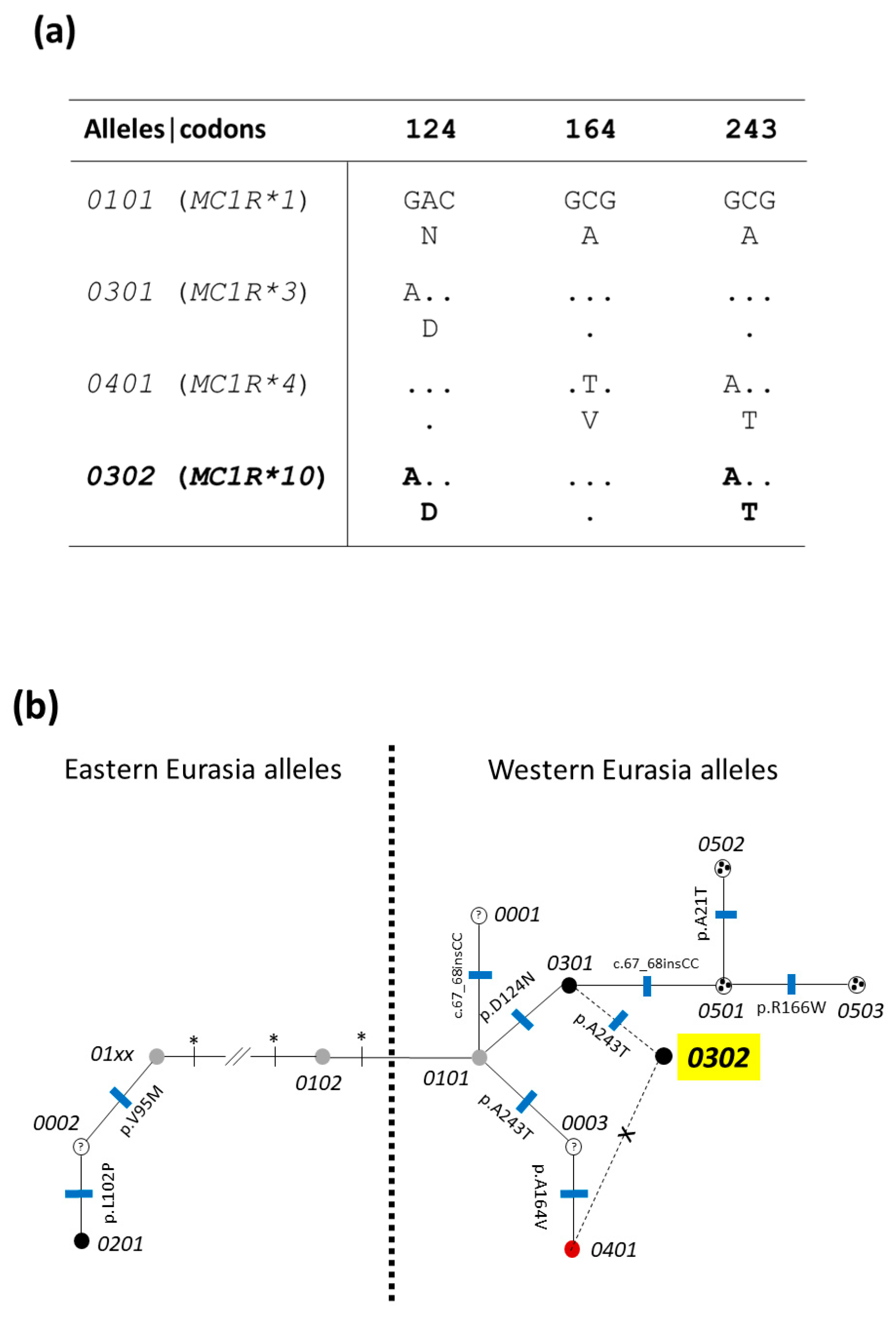
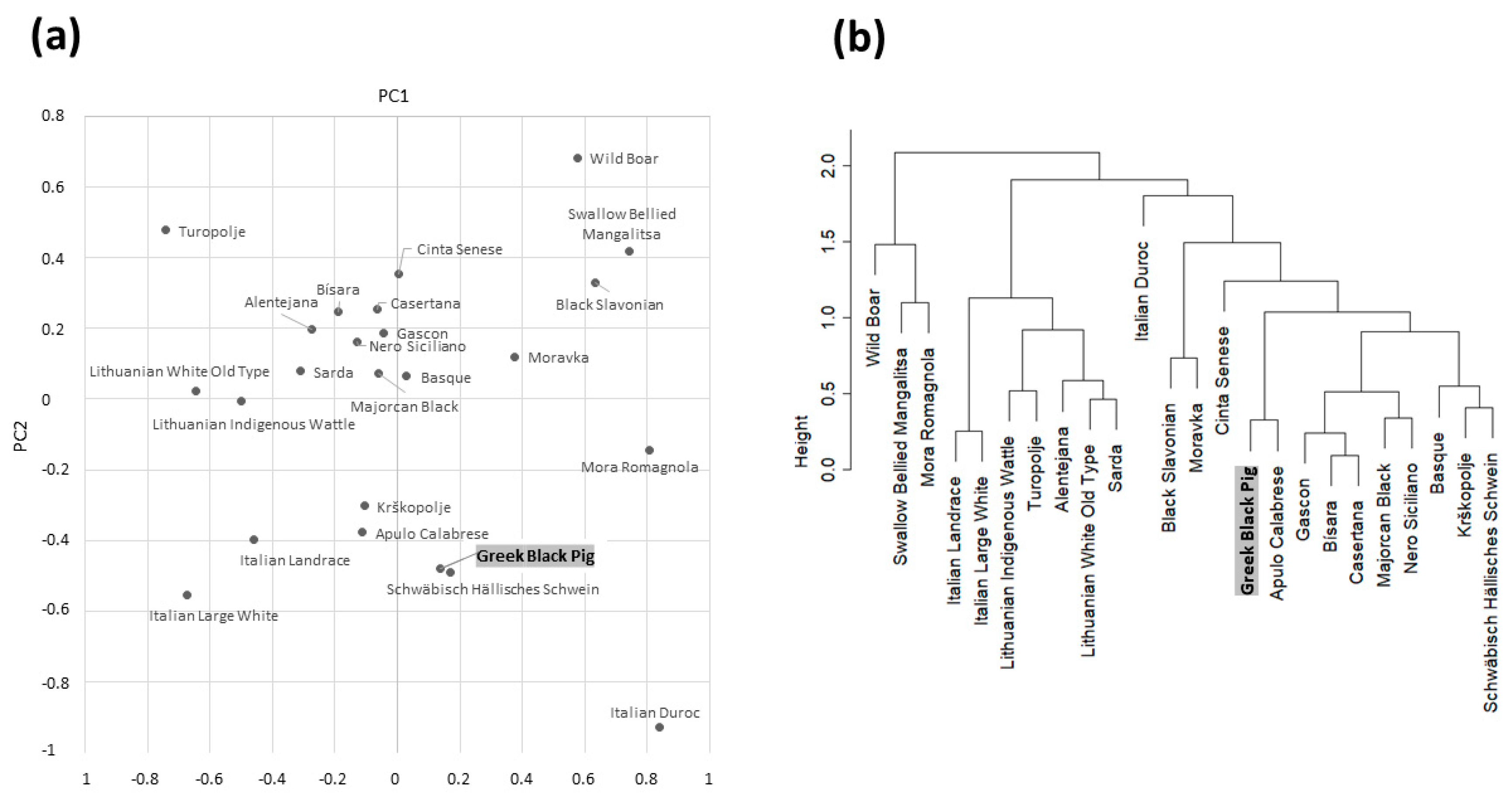
| Gene | MC1R | KIT 1 | NR6A1 2 | VRTN 2 | IGF2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | E+ | ED1 | ED2 | ED2e | EP | E | C | T | T | C | Q | wt | G | A |
| Frequency | 0.076 | 0.009 | 0.703 | 0.042 | 0.017 | 0.153 | 0.983 | 0.017 | 0.161 | 0.839 | 0.449 | 0.551 | 0.144 | 0.856 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribani, A.; Taurisano, V.; Karatosidi, D.; Schiavo, G.; Bovo, S.; Bertolini, F.; Fontanesi, L. Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits. Animals 2023, 13, 1763. https://doi.org/10.3390/ani13111763
Ribani A, Taurisano V, Karatosidi D, Schiavo G, Bovo S, Bertolini F, Fontanesi L. Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits. Animals. 2023; 13(11):1763. https://doi.org/10.3390/ani13111763
Chicago/Turabian StyleRibani, Anisa, Valeria Taurisano, Despoina Karatosidi, Giuseppina Schiavo, Samuele Bovo, Francesca Bertolini, and Luca Fontanesi. 2023. "Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits" Animals 13, no. 11: 1763. https://doi.org/10.3390/ani13111763
APA StyleRibani, A., Taurisano, V., Karatosidi, D., Schiavo, G., Bovo, S., Bertolini, F., & Fontanesi, L. (2023). Signatures of Admixture and Genetic Uniqueness in the Autochthonous Greek Black Pig Breed Deduced from Gene Polymorphisms Affecting Domestication-Derived Traits. Animals, 13(11), 1763. https://doi.org/10.3390/ani13111763







