An Outbreak of Limping Syndrome Associated with Feline Calicivirus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Cases and Sample Collection
2.2. Nucleic Acid Extraction from Samples
2.3. Molecular Screening for FCV
2.4. Molecular Screening for Other Pathogens
2.5. Cells and Virus
2.6. Virus Isolation
2.7. Viral Titration
2.8. Seroneutralization (SN)
2.9. Evaluation of Susceptibility to pH, Trypsin, and Bile Salts
2.10. Full-Genome Amplification
2.11. Oxford Nanopore Sequencing
2.12. Sequence and Phylogenetic Analysis
2.13. Analysis of the Hypervariable Region E
2.14. GenBank Sequence Submission
3. Results
3.1. Molecular Investigation, Virus Isolation, and Titration
3.2. Sequence Analysis of FCV
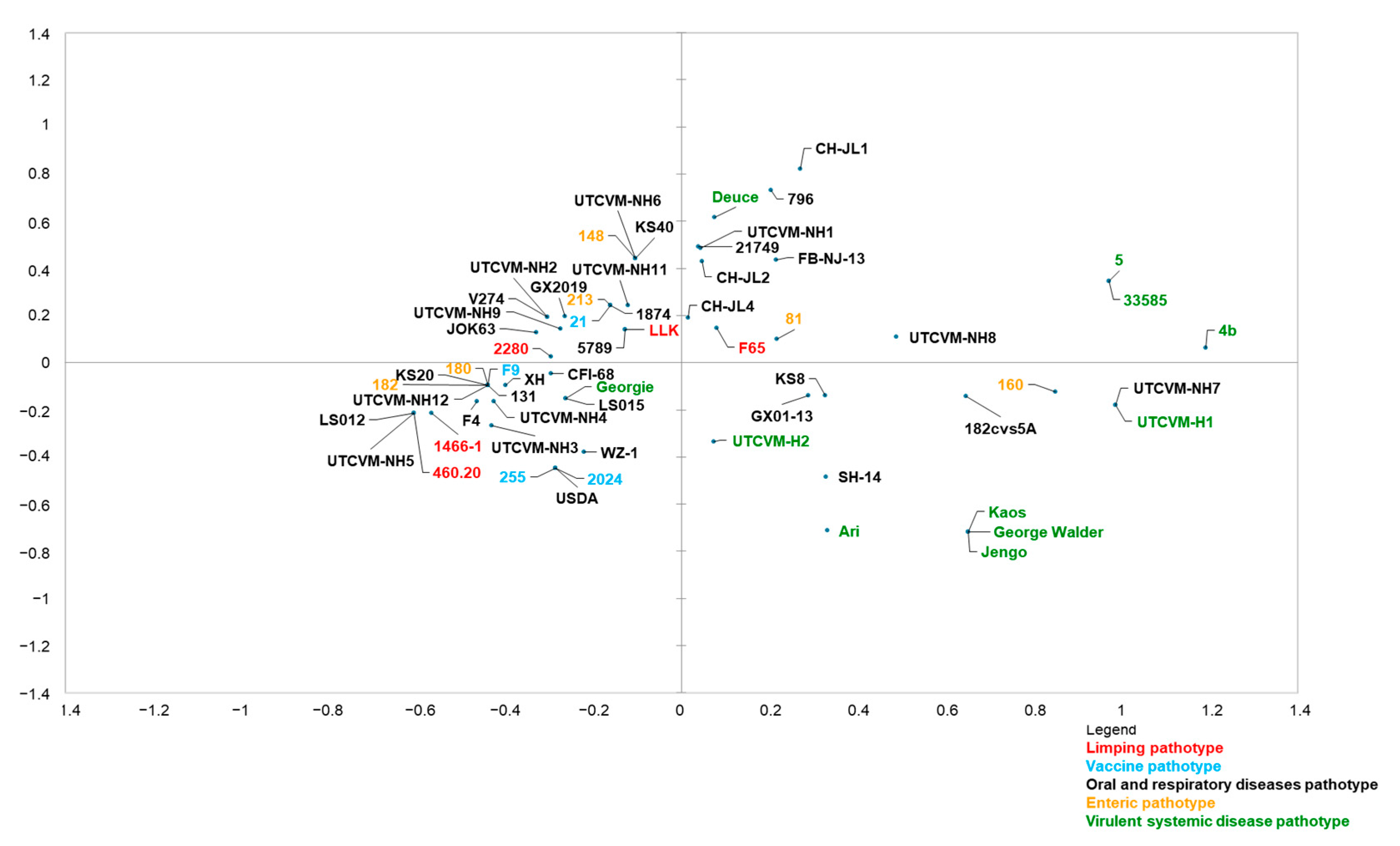
3.3. MCA
3.4. SN
3.5. Evaluation of Susceptibility to pH, Trypsin, and Bile Salts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vinjé, J.; Estes, M.K.; Esteves, P.; Green, K.Y.; Katayama, K.; Knowles, N.J.; L’Homme, Y.; Martella, V.; Vennema, H.; White, P.A. ICTV Virus Taxonomy Profile: Caliciviridae. J. Gen. Virol. 2019, 100, 1469–1470. [Google Scholar] [CrossRef]
- Green, C.; Chalker, V. Immunoprophylaxis. In Infectious Diseases of the Dog and Cat; Saunders Elsevier: St. Louis, MO, USA, 2012; pp. 1163–1205. [Google Scholar]
- Seal, B.S.; Ridpath, J.F.; Mengeling, W.L. Analysis of Feline Calicivirus Capsid Protein Genes: Identification of Variable Antigenic Determinant Regions of the Protein. J. Gen. Virol. 1993, 74, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtsev, S.V.; Green, K.Y. Identification and Genomic Mapping of the ORF3 and VPg Proteins in Feline Calicivirus Virions. Virology 2000, 277, 193–203. [Google Scholar] [CrossRef]
- Radford, A.D.; Addie, D.; Belák, S.; Boucraut-Baralon, C.; Egberink, H.; Frymus, T.; Gruffydd-Jones, T.; Hartmann, K.; Hosie, M.J.; Lloret, A.; et al. Feline Calicivirus Infection: ABCD Guidelines on Prevention and Management. J. Feline Med. Surg. 2009, 11, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Willi, B.; Meli, M.L.; Boretti, F.S.; Hartnack, S.; Dreyfus, A.; Lutz, H.; Hofmann-Lehmann, R. Feline Calicivirus and Other Respiratory Pathogens in Cats with Feline Calicivirus-Related Symptoms and in Clinically Healthy Cats in Switzerland. BMC Vet. Res. 2015, 11, 282. [Google Scholar] [CrossRef] [PubMed]
- Coyne, K.P.; Christley, R.M.; Pybus, O.G.; Dawson, S.; Gaskell, R.M.; Radford, A.D. Large-Scale Spatial and Temporal Genetic Diversity of Feline Calicivirus. J. Virol. 2012, 86, 11356–11367. [Google Scholar] [CrossRef]
- Seal, B.S.; Neill, J.D. Capsid Protein Gene Sequence of Feline Calicivirus Isolates 255 and LLK: Further Evidence for Capsid Protein Configuration among Feline Caliciviruses. Virus Genes 1995, 9, 183–187. [Google Scholar] [CrossRef]
- Hofmann-Lehmann, R.; Hosie, M.J.; Hartmann, K.; Egberink, H.; Truyen, U.; Tasker, S.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A.; et al. Calicivirus Infection in Cats. Viruses 2022, 14, 937. [Google Scholar] [CrossRef]
- Wensman, J.J.; Samman, A.; Lindhe, A.; Thibault, J.-C.; Berndtsson, L.T.; Hosie, M.J. Ability of Vaccine Strain Induced Antibodies to Neutralize Field Isolates of Caliciviruses from Swedish Cats. Acta Vet. Scand. 2015, 57, 86. [Google Scholar] [CrossRef]
- Bergmann; Speck; Rieger; Truyen; Hartmann Antibody Response to Feline Calicivirus Vaccination in Healthy Adult Cats. Viruses 2019, 11, 702. [CrossRef]
- Dawson, S.; Bennett, D.; Carter, S.D.; Bennett, M.; Meanger, J.; Turner, P.C.; Carter, M.J.; Milton, I.; Gaskell, R.M. Acute Arthritis of Cats Associated with Feline Calicivirus Infection. Res. Vet. Sci. 1994, 56, 133–143. [Google Scholar] [CrossRef] [PubMed]
- TerWee, J.; Lauritzen, A.Y.; Sabara, M.; Dreier, K.J.; Kokjohn, K. Comparison of the Primary Signs Induced by Experimental Exposure to Either a Pneumotrophic or a ‘Limping’ Strain of Feline Calicivirus. Vet. Microbiol. 1997, 56, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.; Laliberte, L.; Ekman, S. A Transient Febrile Limping Syndrome of Kittens Caused by Two Different Strains of Feline Calicivirus. Feline Pract. 1983, 13, 26–36. [Google Scholar]
- Bennett, D.; Gaskell, R.; Mills, A.; Knowles, J.; Carter, S.; McArdle, F. Detection of Feline Calicivirus Antigens in the Joints of Infected Cats. Vet. Rec. 1989, 124, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Meli, M.L.; Berger, A.; Willi, B.; Spiri, A.M.; Riond, B.; Hofmann-Lehmann, R. Molecular Detection of Feline Calicivirus in Clinical Samples: A Study Comparing Its Detection by RT-QPCR Directly from Swabs and after Virus Isolation. J. Virol. Methods 2018, 251, 54–60. [Google Scholar] [CrossRef]
- Marsilio, F.; Di Martino, B.; Decaro, N.; Buonavoglia, C. A Novel Nested PCR for the Diagnosis of Calicivirus Infections in the Cat. Vet. Microbiol. 2005, 105, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vogtlin, A.; Fraefel, C.; Albini, S.; Leutenegger, C.M.; Schraner, E.; Spiess, B.; Lutz, H.; Ackermann, M. Quantification of Feline Herpesvirus 1 DNA in Ocular Fluid Samples of Clinically Diseased Cats by Real-Time TaqMan PCR. J. Clin. Microbiol. 2002, 40, 519–523. [Google Scholar] [CrossRef]
- Quackenbush, S.L.; Dean, G.A.; Mullins, J.I.; Hoover, E.A. Analysis of FeLV-FAIDS Provirus Burden and Productive Infection in Lymphocyte Subsets In Vivo. Virology 1996, 223, 1–9. [Google Scholar] [CrossRef]
- Morton, J.M.; McCoy, R.J.; Kann, R.K.C.; Gardner, I.A.; Meers, J. Validation of Real-Time Polymerase Chain Reaction Tests for Diagnosing Feline Immunodeficiency Virus Infection in Domestic Cats Using Bayesian Latent Class Models. Prev. Vet. Med. 2012, 104, 136–148. [Google Scholar] [CrossRef]
- Di Martino, B.; Lanave, G.; Di Profio, F.; Melegari, I.; Marsilio, F.; Camero, M.; Catella, C.; Capozza, P.; Bányai, K.; Barrs, V.R.; et al. Identification of Feline Calicivirus in Cats with Enteritis. Transbound. Emerg. Dis. 2020, 67, 2579–2588. [Google Scholar] [CrossRef]
- Brunet, S.; Sigoillot-Claude, C.; Pialot, D.; Poulet, H. Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains. Viruses 2019, 11, 1090. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Verin, R.; Buldrini, I.; Zamagni, S.; Morini, M.; Terrusi, A.; Gallina, L.; Urbani, L.; Dondi, F.; Battilani, M. Natural Cases of Polyarthritis Associated with Feline Calicivirus Infection in Cats. Vet. Res. Commun. 2022, 46, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Spiri, A.M. An Update on Feline Calicivirus. Schweiz. Arch. Tierheilkd. 2022, 164, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Ossiboff, R.J.; Sheh, A.; Shotton, J.; Pesavento, P.A.; Parker, J.S.L. Feline Caliciviruses (FCVs) Isolated from Cats with Virulent Systemic Disease Possess in Vitro Phenotypes Distinct from Those of Other FCV Isolates. J. Gen. Virol. 2007, 88, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Bordicchia, M.; Fumian, T.M.; Van Brussel, K.; Russo, A.G.; Carrai, M.; Le, S.-J.; Pesavento, P.A.; Holmes, E.C.; Martella, V.; White, P.; et al. Feline Calicivirus Virulent Systemic Disease: Clinical Epidemiology, Analysis of Viral Isolates and In Vitro Efficacy of Novel Antivirals in Australian Outbreaks. Viruses 2021, 13, 2040. [Google Scholar] [CrossRef]
- Povey, R.C.; Hale, C.J. Experimental Infections with Feline Caliciviruses (Picornaviruses) in Specific-Pathogen-Free Kittens. J. Comp. Pathol. 1974, 84, 245–256. [Google Scholar] [CrossRef]
- Mochizuki, M. Different Stabilities to Bile among Feline Calicivirus Strains of Respiratory and Enteric Origin. Vet. Microbiol. 1992, 31, 297–302. [Google Scholar] [CrossRef]
- Geissler, K.; Schneider, K.; Platzer, G.; Truyen, B.; Kaaden, O.-R.; Truyen, U. Genetic and Antigenic Heterogeneity among Feline Calicivirus Isolates from Distinct Disease Manifestations. Virus Res. 1997, 48, 193–206. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Elliott, J.B.; Glasgow, A.; Poland, A.; Keel, K. An Isolated Epizootic of Hemorrhagic-like Fever in Cats Caused by a Novel and Highly Virulent Strain of Feline Calicivirus. Vet. Microbiol. 2000, 73, 281–300. [Google Scholar] [CrossRef]
- Poulet, H.; Brunet, S.; Soulier, M.; Leroy, V.; Goutebroze, S.; Chappuis, G. Comparison between Acute Oral/Respiratory and Chronic Stomatitis/Gingivitis Isolates of Feline Calicivirus: Pathogenicity, Antigenic Profile and Cross-Neutralisation Studies. Arch. Virol. 2000, 145, 243–261. [Google Scholar] [CrossRef]
- Pedersen, N.; Pool, R.; Morgan, J. Diseases of the Dog and Cat. In Textbook of Verterinary Internal Medicine; Ettinger: Philadelphia, PA, USA, 1983. [Google Scholar]
- Church, R.E. Lameness in Kittens after Vaccination. Vet. Rec. 1989, 125, 609. [Google Scholar]
- Levy, J.K.; Marsh, A. Isolation of Calicivirus from the Joint of a Kitten with Arthritis. J. Am. Vet. Med. Assoc. 1992, 201, 753–755. [Google Scholar]
- Dawson, S.; McArdle, F.; Bennett, M.; Carter, M.; Milton, I.; Turner, P.; Meanger, J.; Gaskell, R. Typing of Feline Calicivirus Isolates from Different Clinical Groups by Virus Neutralisation Tests. Vet. Rec. 1993, 133, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Hurley, K.F.; Sykes, J.E. Update on Feline Calicivirus: New Trends. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Doultree, J.C.; Druce, J.D.; Birch, C.J.; Bowden, D.S.; Marshall, J.A. Inactivation of Feline Calicivirus, a Norwalk Virus Surrogate. J. Hosp. Infect. 1999, 41, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.D.; Coyne, K.P.; Dawson, S.; Porter, C.J.; Gaskell, R.M. Feline Calicivirus. Vet. Res. 2007, 38, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Mackiewicz, A.; Pawo̵wski, T.; Mackiewicz-Pawłowska, A.; Wiktorowicz, K.; Mackiewicz, S. Microheterogeneity Forms of Alpha1-Acid Glycoprotein as Indicators of Rheumatoid Arthritis Activity. Clin. Chim. Acta 1987, 163, 185–190. [Google Scholar] [CrossRef]
- Tian, J.; Kang, H.; Huang, J.; Li, Z.; Pan, Y.; Li, Y.; Chen, S.; Zhang, J.; Yin, H.; Qu, L. Feline Calicivirus Strain 2280 P30 Antagonizes Type I Interferon-Mediated Antiviral Innate Immunity through Directly Degrading IFNAR1 MRNA. PLoS Pathog. 2020, 16, e1008944. [Google Scholar] [CrossRef] [PubMed]
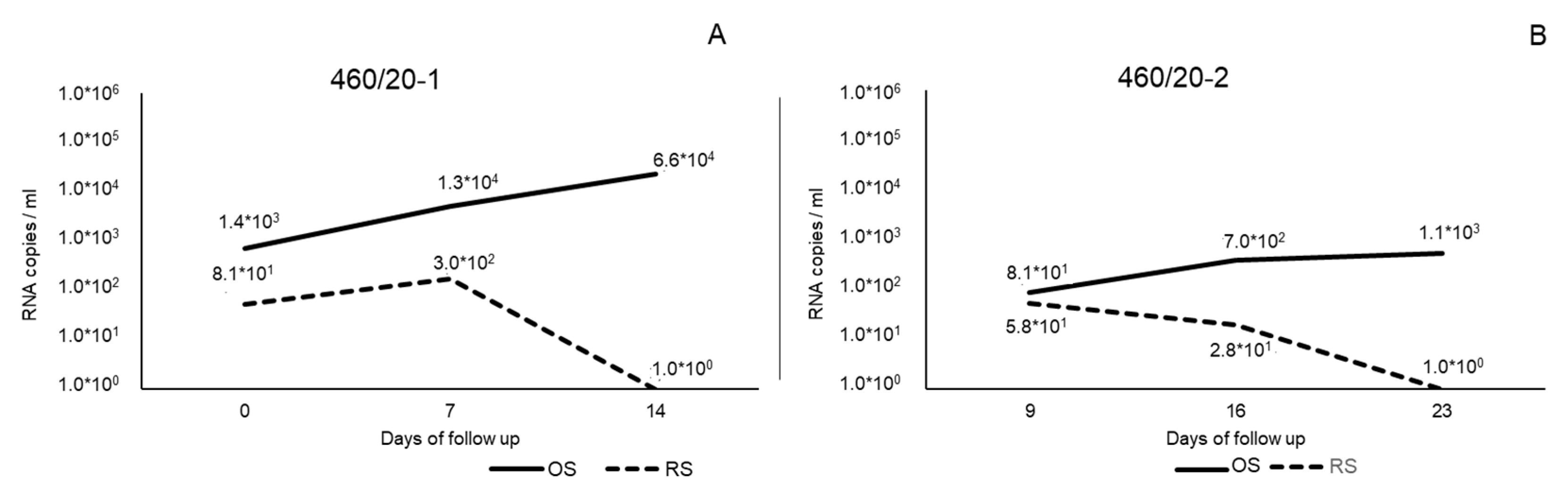


| Cat | Clinical Signs | Days of Follow Up | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | ||
| 460/20-1 | Lameness | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | - | - | - | - | - | - | - | - | - | - |
| Fever | - | - | • | • | • | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Oral lesions | - | - | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | - | - | - | - | - | - | - | - | - | - | |
| Respiratory signs | - | • | • | • | • | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Oral swab | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | |
| Rectal swab | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | |
| Blood and serum | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Hospitalization | - | - | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | - | - | - | - | - | - | - | - | - | - | |
| 460/20-2 | Lameness | - | - | - | - | - | - | - | - | - | - | - | • | • | • | • | • | • | • | • | • | • | • | - | - | - | - | - |
| Fever | - | - | - | - | - | - | - | - | - | - | - | • | • | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Oral lesions | - | - | - | - | - | - | - | - | - | - | - | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | |
| Respiratory signs | - | - | - | - | - | - | - | - | - | - | - | • | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Oral swab | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | |
| Rectal swab | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | - | - | - | - | - | T | - | |
| Blood and serum | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | |
| Hospitalization | - | - | - | - | - | - | - | - | - | - | - | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | ▲ | |
| Strain | Pathotype | Country | Year | Access Number |
|---|---|---|---|---|
| 1466-1 | LIMP | Italy | 2017 | MT062980 |
| 2280 | LIMP | Canada | 1982 | KC835209 |
| F65 | LIMP | United Kingdom | 1990 | AF109465 |
| LLK | LIMP | Canada | 1982 | U07131 |
| 21 | VAC | USA | 1993 | KJ021044 |
| 255 | VAC | USA | 1970 | KM111170 |
| 2024 | VAC | Germany | 2002 | AF479590 |
| F9 | VAC | USA | 1958 | M86379 |
| 131 | ORD | USA | 2003 | DQ910787 |
| 1874 | ORD | USA | 1996 | JX519214 |
| CFI-68 | ORD | USA | 1968 | U13992 |
| CH-JL1 | ORD | China | 2013 | KJ495728 |
| CH-JL2 | ORD | China | 2013 | KJ495729 |
| CH-JL4 | ORD | China | 2015 | KT206207 |
| F4 | ORD | Japan | 1971 | D31836 |
| FB-NJ-13 | ORD | China | 2013 | KM111557 |
| GX01-13 | ORD | China | 2013 | KT970059 |
| GX2019 | ORD | China | 2019 | MK867378 |
| JOK63 | ORD | United Kingdom | 1985 | AF109466 |
| KS40 | ORD | Germany | 1995 | X99448 |
| SH-14 | ORD | China | 2014 | KT000003 |
| USDA | ORD | USA | 2005 | AY560118 |
| UTCVM-NH2 | ORD | USA | 1998 | AY560114 |
| UTCVM-NH3 | ORD | USA | 2000 | AY560115 |
| UTCVM-NH4 | ORD | USA | 2009 | AY996855 |
| UTCVM-NH5 | ORD | USA | 2009 | AY996856 |
| UTCVM-NH6 | ORD | USA | 2009 | AY996857 |
| UTCVM-NH7 | ORD | USA | 2009 | AY996858 |
| UTCVM-NH8 | ORD | USA | 2009 | AY996859 |
| UTCVM-NH9 | ORD | USA | 2009 | AY996860 |
| UTCVM-NH11 | ORD | USA | 2009 | AY996862 |
| UTCVM-NH12 | ORD | USA | 2009 | AY996863 |
| V274 | ORD | Australia | 1989 | AF031877 |
| WZ-1 | ORD | China | 2016 | KX371573 |
| XH | ORD | China | 2015 | KX371572 |
| 182cvs5A | ORD | Australia | 1980 | AF031875 |
| 796 | ORD | USA | 2004 | DQ910788 |
| 5789 | ORD | USA | 1996 | JX519210 |
| 21749 | ORD | USA | 1996 | JX519213 |
| KS8 | ORD | Germany | 1994 | X99449 |
| KS20 | ORD | Germany | 1994 | X99447 |
| LS012 | ORD | United Kingdom | 1985 | AF109467 |
| LS015 | ORD | United Kingdom | 1985 | AF109464 |
| UTCVM-NH1 | ORD | USA | 1993 | AY560113 |
| 81 | ENT | Italy | 2015 | MT008246 |
| 148 | ENT | Italy | 2015 | MT008248 |
| 160 | ENT | Italy | 2015 | MT008250 |
| 180 | ENT | Italy | 2015 | MT008249 |
| 182 | ENT | Italy | 2015 | MT008247 |
| 213 | ENT | Italy | 1995 | AF283778 |
| 4b | VSD | France | 2007 | EU202915 |
| 5 | VSD | USA | 2001 | DQ910790 |
| 33585 | VSD | USA | 2000 | EF675759 |
| Ari | VSD | USA | 1998 | DQ910794 |
| Deuce | VSD | USA | 2004 | DQ910789 |
| George Walder | VSD | USA | 2002 | DQ910792 |
| Georgie | VSD | USA | 2001 | DQ910791 |
| Jengo | VSD | USA | 2002 | DQ910793 |
| Kaos | VSD | USA | 2002 | DQ910795 |
| UTCVM-H1 | VSD | USA | 1999 | AY560116 |
| UTCVM-H2 | VSD | USA | 2002 | AY560117 |
| Variable | LIM Strain | p-Value |
|---|---|---|
| 438_Hydrophobic | Y | >0.05 |
| 438_Polar | Y | 0.00325 |
| 438_Small | Y | >0.05 |
| 440_Hydrophobic | N | >0.05 |
| 440_Small | Y | >0.05 |
| 448_Hydrophobic | Y | >0.05 |
| 448_Small | Y | 0.004 |
| 452_Negative | Y | >0.05 |
| 452_Small | Y | 0.0002 |
| 455_Negative | Y | >0.05 |
| 455_Small | Y | >0.05 |
| 465_Hydrophobic | N | 0.0015 |
| 465_Small | Y | >0.05 |
| 492_Positive | N | >0.05 |
| FCV Strain | Log10 Reduction of FCV Titer (log10 TCID50) Resulting from Control Virus and Treatment Indicated | ||
|---|---|---|---|
| HCL (pH 3.0) | Trypsin (0.5%) | Bile (0.5%) | |
| Δ | Δ | Δ | |
| 460.20-1 OS/ITA/2020 | 4.0 | 4.5 | 0.0 |
| 460.20-1-RS/ITA/2020 | 4.0 | 4.0 | 0.0 |
| 460.20-2 OS/ITA/2020 | 4.0 | 3.5 | 0.25 |
| 460.20-2 RS/ITA/2020 | 4.0 | 4.0 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanave, G.; Buonavoglia, A.; Pellegrini, F.; Di Martino, B.; Di Profio, F.; Diakoudi, G.; Catella, C.; Omar, A.H.; Vasinioti, V.I.; Cardone, R.; et al. An Outbreak of Limping Syndrome Associated with Feline Calicivirus. Animals 2023, 13, 1778. https://doi.org/10.3390/ani13111778
Lanave G, Buonavoglia A, Pellegrini F, Di Martino B, Di Profio F, Diakoudi G, Catella C, Omar AH, Vasinioti VI, Cardone R, et al. An Outbreak of Limping Syndrome Associated with Feline Calicivirus. Animals. 2023; 13(11):1778. https://doi.org/10.3390/ani13111778
Chicago/Turabian StyleLanave, Gianvito, Alessio Buonavoglia, Francesco Pellegrini, Barbara Di Martino, Federica Di Profio, Georgia Diakoudi, Cristiana Catella, Ahmed H. Omar, Violetta I. Vasinioti, Roberta Cardone, and et al. 2023. "An Outbreak of Limping Syndrome Associated with Feline Calicivirus" Animals 13, no. 11: 1778. https://doi.org/10.3390/ani13111778





