First Report of Mitochondrial DNA Copy Number Variation in Opsius heydeni (Insecta, Hemiptera, Cicadellidae) from Polluted and Control Sites
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Barcoding
2.3. mtDNAcn
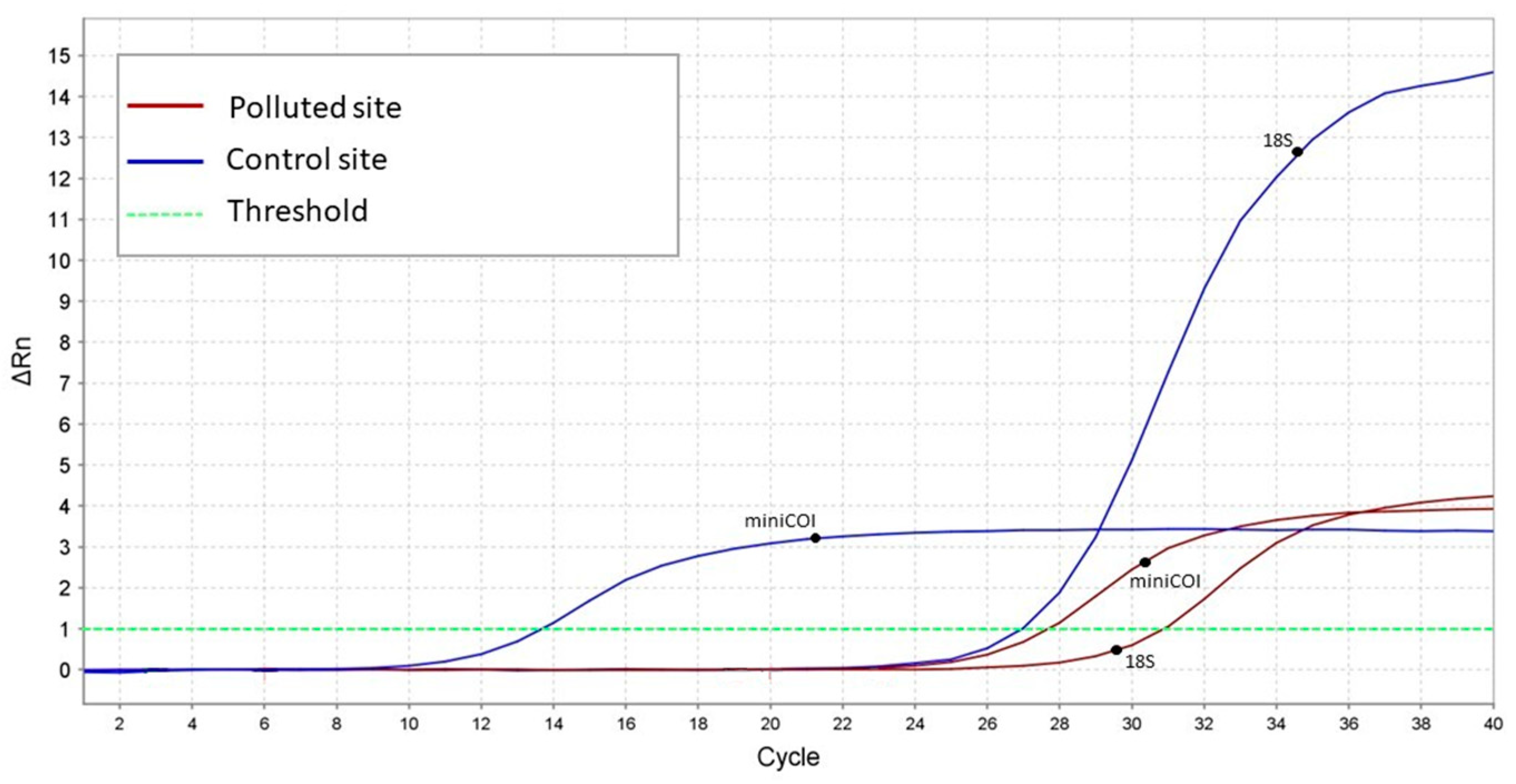
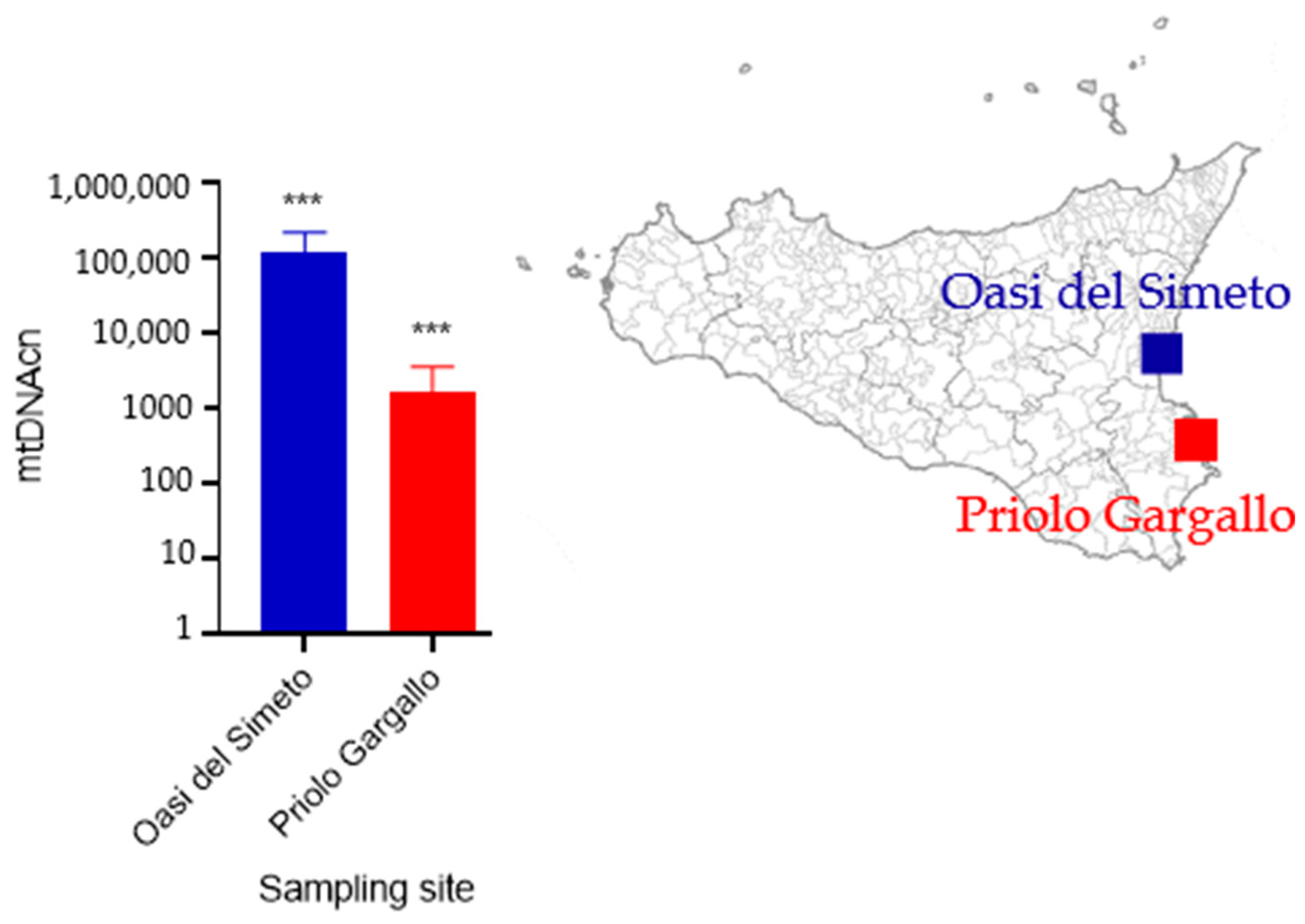
3. Results
3.1. DNA Barcoding
3.2. mtDNAcn Variation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chormare, R.; Kumar, M.A. Environmental Health and Risk Assessment Metrics with Special Mention to Biotransfer, Bioaccumulation and Biomagnification of Environmental Pollutants. Chemosphere 2022, 302, 134836. [Google Scholar] [CrossRef]
- Minh, N.H.; Minh, T.B.; Kajiwara, N.; Kunisue, T.; Iwata, H.; Viet, P.H.; Cam Tu, N.P.; Tuyen, B.C.; Tanabe, S. Pollution Sources and Occurrences of Selected Persistent Organic Pollutants (POPs) in Sediments of the Mekong River Delta, South Vietnam. Chemosphere 2007, 67, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Suedel, B.C.; Boraczek, J.A.; Peddicord, R.K.; Clifford, P.A.; Dillon, T.M. Trophic Transfer and Biomagnification Potential of Contaminants in Aquatic Ecosystems. Rev. Environ. Contam. Toxicol. 1994, 136, 21–89. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.A.; Jolley, D.F.; Simpson, S.L. Effect of Overlying Water PH, Dissolved Oxygen, Salinity and Sediment Disturbances on Metal Release and Sequestration from Metal Contaminated Marine Sediments. Chemosphere 2007, 69, 1428–1437. [Google Scholar] [CrossRef]
- Ibor, O.R.; Eni, G.; Andem, A.B.; Bassey, I.U.; Arong, G.A.; Asor, J.; Regoli, F.; Arukwe, A. Biotransformation and Oxidative Stress Responses in Relation to Tissue Contaminant Burden in Clarias gariepinus Exposed to Simulated Leachate from a Solid Waste Dumpsite in Calabar, Nigeria. Chemosphere 2020, 253, 126630. [Google Scholar] [CrossRef] [PubMed]
- Gredilla, R.; Bohr, V.A.; Stevnsner, T. Mitochondrial DNA Repair and Association with Aging—An Update. Exp. Gerontol. 2010, 45, 478–488. [Google Scholar] [CrossRef]
- Roubicek, D.A.; de Souza-Pinto, N.C. Mitochondria and Mitochondrial DNA as Relevant Targets for Environmental Contaminants. Toxicology 2017, 391, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.N.; Czajka, A. Is Mitochondrial DNA Content a Potential Biomarker of Mitochondrial Dysfunction? Mitochondrion 2013, 13, 481–492. [Google Scholar] [CrossRef]
- Ashar, F.N.; Zhang, Y.; Longchamps, R.J.; Lane, J.; Moes, A.; Grove, M.L.; Mychaleckyj, J.C.; Taylor, K.D.; Coresh, J.; Rotter, J.I.; et al. Association of Mitochondrial DNA Copy Number with Cardiovascular Disease. JAMA Cardiol. 2017, 2, 1247–1255. [Google Scholar] [CrossRef]
- Taylor, R.W.; Turnbull, D.M. Mitochondrial DNA Mutations in Human Disease. Nat. Rev. Genet. 2005, 6, 389–402. [Google Scholar] [CrossRef]
- Fazzini, F.; Lamina, C.; Raftopoulou, A.; Koller, A.; Fuchsberger, C.; Pattaro, C.; Del Greco, F.M.; Döttelmayer, P.; Fendt, L.; Fritz, J.; et al. Association of Mitochondrial DNA Copy Number with Metabolic Syndrome and Type 2 Diabetes in 14 176 Individuals. J. Intern. Med. 2021, 290, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Mutlu, A.G. Increase in Mitochondrial DNA Copy Number in Response to Ochratoxin a and Methanol-Induced Mitochondrial DNA Damage in Drosophila. Bull. Environ. Contam. Toxicol. 2012, 89, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Kim, H.R.; Kang, M.G.; Trang, N.T.D.; Baek, H.J.; Moon, J.D.; Shin, J.H.; Suh, S.P.; Ryang, D.W.; Kook, H.; et al. Profiling of Biomarkers for the Exposure of Polycyclic Aromatic Hydrocarbons: Lamin-A/C Isoform 3, Poly[ADP-Ribose] Polymerase 1, and Mitochondria Copy Number Are Identified as Universal Biomarkers. Biomed Res. Int. 2014, 2014, 605135. [Google Scholar] [CrossRef] [PubMed]
- Law (Italian), No. 426/1998, Nuovi Interventi in Campo Ambientale. Gazzetta Ufficiale 1998; 291. Available online: https://www.gazzettaufficiale.it/eli/gu/2016/07/27/174/sg/pdf(accessed on 27 May 2023).
- Martuzzi, M.; Mitis, F.; Biggeri, A.; Terracini, B.; Bertollini, R. Ambiente e stato di salute nella popolazione delle aree ad alto rischio di crisi ambientale in Italia. Epidemiol Prev. 2002, 26, 1–53. [Google Scholar]
- Di Bella, C.; Traina, A.; Giosuè, C.; Carpintieri, D.; Lo Dico, G.M.; Bellante, A.; Del Core, M.; Falco, F.; Gherardi, S.; Uccello, M.M.; et al. Heavy Metals and PAHs in Meat, Milk, and Seafood from Augusta Area (Southern Italy): Contamination Levels, Dietary Intake, and Human Exposure Assessment. Front. Public Health 2020, 8, 273. [Google Scholar] [CrossRef]
- Benedetti, M.; Romano, E.; Ausili, A.; Fattorini, D.; Gorbi, S.; Maggi, C.; Salmeri, A.; Salvagio Manta, D.; Sesta, G.; Sprovieri, M.; et al. 10-Year Time Course of Hg and Organic Compounds in Augusta Bay: Bioavailability and Biological Effects in Marine Organisms. Front. Public Health 2022, 10, 968296. [Google Scholar] [CrossRef]
- Maisano, M.; Cappello, T.; Natalotto, A.; Vitale, V.; Parrino, V.; Giannetto, A.; Oliva, S.; Mancini, G.; Cappello, S.; Mauceri, A.; et al. Effects of Petrochemical Contamination on Caged Marine Mussels Using a Multi-Biomarker Approach: Histological Changes, Neurotoxicity and Hypoxic Stress. Mar. Environ. Res. 2017, 128, 114–123. [Google Scholar] [CrossRef]
- Copat, C.; Ferrante, M.; Hernout, B.V.; Giunta, F.; Grasso, A.; Messina, A.; Grasso, R.; Spena, M.T. Trace Element Bioaccumulation in Stone Curlew (Burhinus oedicnemus, Linnaeus, 1758): A Case Study from Sicily (Italy). Int. J. Mol. Sci. 2020, 21, 4597. [Google Scholar] [CrossRef]
- El-Sonbati, S.A.; Wilson, M.R.; Al Dhafer, H.M. The Tamarix Feeding Leafhopper Genus Opsius Fieber, 1866 (Hemiptera, Cicadellidae, Deltocephalinae, Opsiini) in the Kingdom of Saudi Arabia, with Description of a New Species. Dtsch. Entomol. Z. 2020, 67, 1–12. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The Number of Known Plants Species in the World and Its Annual Increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Fawzy, E.M.; Soltan, M.E.; Sirry, S.M. Mobilization of Different Metals between Tamarix Parts and Their Crystal Salts-Soil System at the Banks of River Nile, Aswan, Egypt. Toxicol. Environ. Chem. 2006, 88, 603–618. [Google Scholar] [CrossRef]
- Manousaki, E.; Kadukova, J.; Papadantonakis, N.; Kalogerakis, N. Phytoextraction and Phytoexcretion of Cd by the Leaves of Tamarix smyrnensis Growing on Contaminated Non-Saline and Saline Soils. Environ. Res. 2008, 106, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Jeddi, K.; Fatnassi, M.; Chaieb, M.; Siddique, K.H.M. Tree Species as a Biomonitor of Metal Pollution in Arid Mediterranean Environments: Case for Arid Southern Tunisia. Environ. Sci. Pollut. Res. 2021, 28, 28598–28605. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D.G.M.; Viscuso, R.; D’Urso, V.; Gibilras, S.; Sardella, A.; Marletta, A.; Pappalardo, A.M. Morphostructural Analysis of the Male Reproductive System and DNA Barcoding in Balclutha brevis Lindberg 1954 (Homoptera, Cicadellidae). Micron 2015, 79, 36–45. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Wittig, I.; Dröse, S.; Schmeisser, S.; Lück, C.; Hahn, C.; Graf, M.; Gausmann, U.; Terzibasi, E.; et al. Mitochondrial DNA Copy Number and Function Decrease with Age in the Short-Lived Fish Nothobranchius furzeri. Aging Cell 2011, 10, 824–831. [Google Scholar] [CrossRef]
- Messina, C.M.; Arena, R.; Manuguerra, S.; La Barbera, L.; Curcuraci, E.; Renda, G.; Santulli, A. Valorization of Side Stream Products from Sea Cage Fattened Bluefin Tuna (Thunnus thynnus): Production and In Vitro Bioactivity Evaluation of Enriched ω-3 Polyunsaturated Fatty Acids. Mar. Drugs 2022, 20, 309. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Loeschcke, V.; Tan, Q.; Pertoldi, C.; Mengel-From, J. Sex and Age Specific Reduction in Stress Resistance and Mitochondrial DNA Copy Number in Drosophila melanogaster. Sci. Rep. 2019, 9, 12305. [Google Scholar] [CrossRef]
- Li, Z.H.; Zhang, P.; Ma, H.K.; Xu, W.Y.; Sun, J.Q.; Yan, B.L.; Zhang, Q.Q.; Gao, H. Effect of Temperature and Salinity on MtDNA Copy Number of the Ridgetail White Prawn, Palaemon carinicauda Holthuis, 1950 (Decapoda, Palaemonidae). Crustaceana 2018, 91, 1061–1072. [Google Scholar] [CrossRef]
- Syromyatnikov, M.Y.; Gureev, A.P.; Mikhaylov, E.V.; Parshin, P.A.; Popov, V.N. Pesticides Effect on the Level of MtDNA Damage in Bumblebees Heads (Bombus terrestris L.). Period. Tche Quim. 2020, 17, 395–402. [Google Scholar] [CrossRef]
- Li, Z.; Su, Q.; Xu, R.; Peng, J.; Zhu, X.; Wei, Y. Influence of Different Concentrations of Ozone on the Alteration of Mitochondrial DNA Copy Numbers in Human Peripheral Blood. Sci. Total Environ. 2023, 873, 162282. [Google Scholar] [CrossRef] [PubMed]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA Copy Number in Human Disease: The More the Better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.A.; Coombs, M.M. Covalent Binding of Polycyclic Aromatic Compounds to Mitochondrial and Nuclear DNA. Nature 1980, 287, 244–245. [Google Scholar] [CrossRef]
- Pavanello, S.; Dioni, L.; Hoxha, M.; Fedeli, U.; Mielzynska-Švach, D.; Baccarelli, A.A. Mitochondrial Dna Copy Number and Exposure to Polycyclic Aromatic Hydrocarbons. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1722–1729. [Google Scholar] [CrossRef]
- Carugno, M.; Pesatori, A.C.; Dioni, L.; Hoxha, M.; Bollati, V.; Albetti, B.; Byun, H.M.; Bonzini, M.; Fustinoni, S.; Cocco, P.; et al. Increased Mitochondrial DNA Copy Number in Occupations Associated with Low-Dose Benzene Exposure. Environ. Health Perspect. 2012, 120, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Chinde, S.; Kumari, M.; Devi, K.R.; Murty, U.S.; Rahman, M.F.; Kumari, S.I.; Mahboob, M.; Grover, P. Assessment of Genotoxic Effects of Lead in Occupationally Exposed Workers. Environ. Sci. Pollut. Res. 2014, 21, 11469–11480. [Google Scholar] [CrossRef]
- Xu, Y.; Lindh, C.H.; Jönsson, B.A.G.; Broberg, K.; Albin, M. Occupational Exposure to Asphalt Mixture during Road Paving Is Related to Increased Mitochondria DNA Copy Number: A Cross-Sectional Study. Environ. Health A Glob. Access Sci. Source 2018, 17, 29. [Google Scholar] [CrossRef]
- Gerlach, J.; Samways, M.; Pryke, J. Terrestrial Invertebrates as Bioindicators: An Overview of Available Taxonomic Groups. J. Insect Conserv. 2013, 17, 831–850. [Google Scholar] [CrossRef]

| Sample Code | GenBank | Species Matched by BLAST | Matched GenBank | % Identity |
|---|---|---|---|---|
| Accession N° | Accession from BLAST | 100% Coverage | ||
| Oh1 | OQ661886 | Opsius stactogalus | HQ929190 | 83.66 |
| Oh2 | OQ661887 | Opsius stactogalus | HQ929190 | 84.12 |
| Oh3 | OQ661888 | Opsius stactogalus | HQ929190 | 83.97 |
| Oh4 | OQ661889 | Opsius stactogalus | HQ929190 | 83.51 |
| Oh5 | OQ661890 | Opsius stactogalus | HQ929190 | 83.66 |
| Oh6 | OQ661891 | Opsius stactogalus | HQ929190 | 83.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calogero, G.S.; Giuga, M.; D’Urso, V.; Ferrito, V.; Pappalardo, A.M. First Report of Mitochondrial DNA Copy Number Variation in Opsius heydeni (Insecta, Hemiptera, Cicadellidae) from Polluted and Control Sites. Animals 2023, 13, 1793. https://doi.org/10.3390/ani13111793
Calogero GS, Giuga M, D’Urso V, Ferrito V, Pappalardo AM. First Report of Mitochondrial DNA Copy Number Variation in Opsius heydeni (Insecta, Hemiptera, Cicadellidae) from Polluted and Control Sites. Animals. 2023; 13(11):1793. https://doi.org/10.3390/ani13111793
Chicago/Turabian StyleCalogero, Giada Santa, Marta Giuga, Vera D’Urso, Venera Ferrito, and Anna Maria Pappalardo. 2023. "First Report of Mitochondrial DNA Copy Number Variation in Opsius heydeni (Insecta, Hemiptera, Cicadellidae) from Polluted and Control Sites" Animals 13, no. 11: 1793. https://doi.org/10.3390/ani13111793





