SINE Insertion in the Pig Carbonic Anhydrase 5B (CA5B) Gene Is Associated with Changes in Gene Expression and Phenotypic Variation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. DNA Isolation and Amplification
2.3. RIP Verification and Genotyping by PCR
2.4. Economic Trait Data Collection
2.5. Expression Analysis
2.6. Vector Construction
2.7. Dual-Luciferase Reporter Assay
2.8. Cloning and Sequencing of the SINE-Carried (ATTT)n Repeat Sequence across Genomes
2.9. Statistical Analysis and Growth Correlation Analysis
3. Results
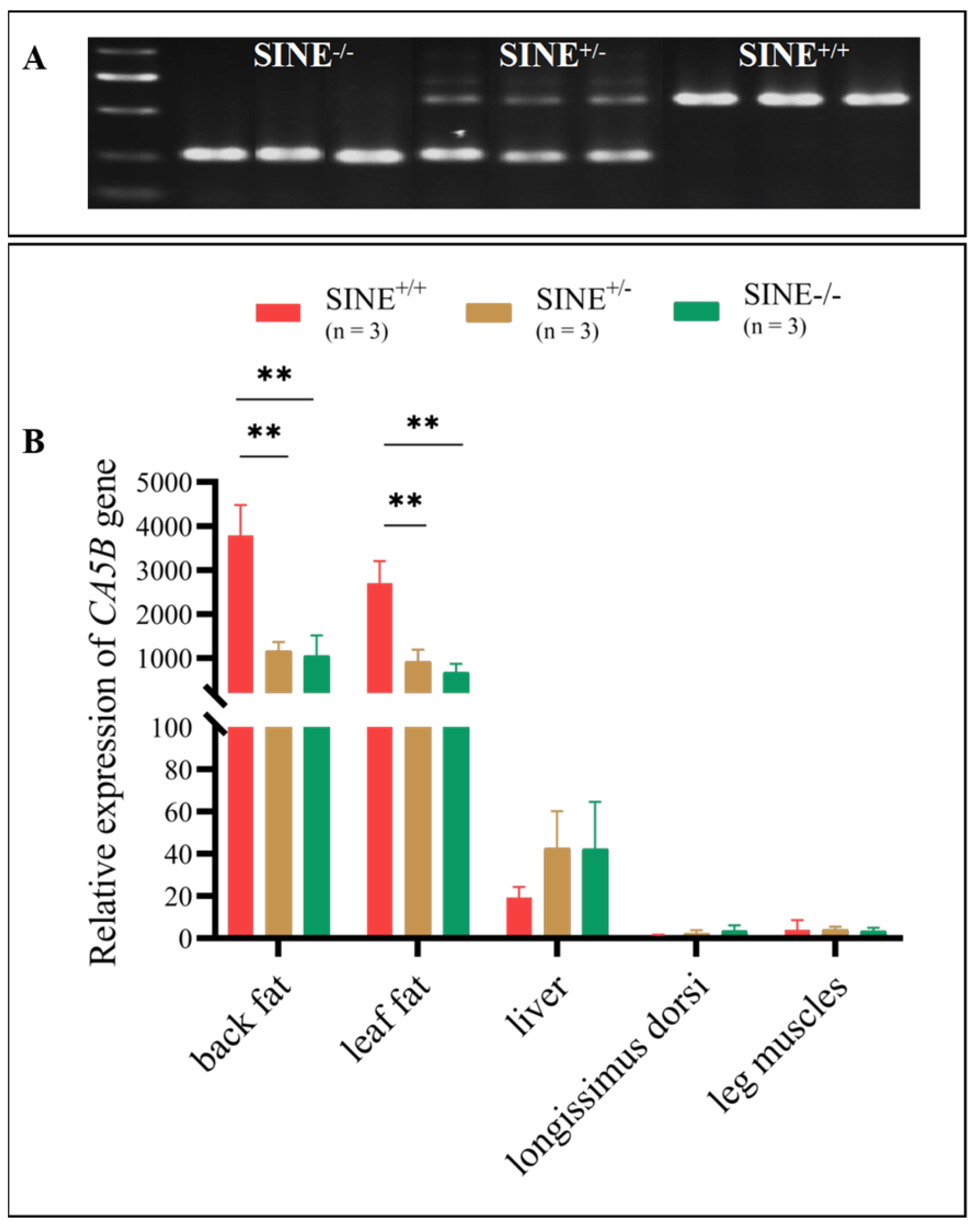
3.1. SINE RIP in the First Intron of Pig CA5B Gene Verified by PCR and Sequencing
3.2. RIP Distribution in Different Pig Breeds
3.3. Increased CA5B Expression in Fat Tissues Associated with the SINE-RIP in the First Intron of the CA5B Gene
3.4. Impact of SINE Insertion in the First Intron of CA5B Gene on Regulatory Activity
3.5. Growth Association Analysis
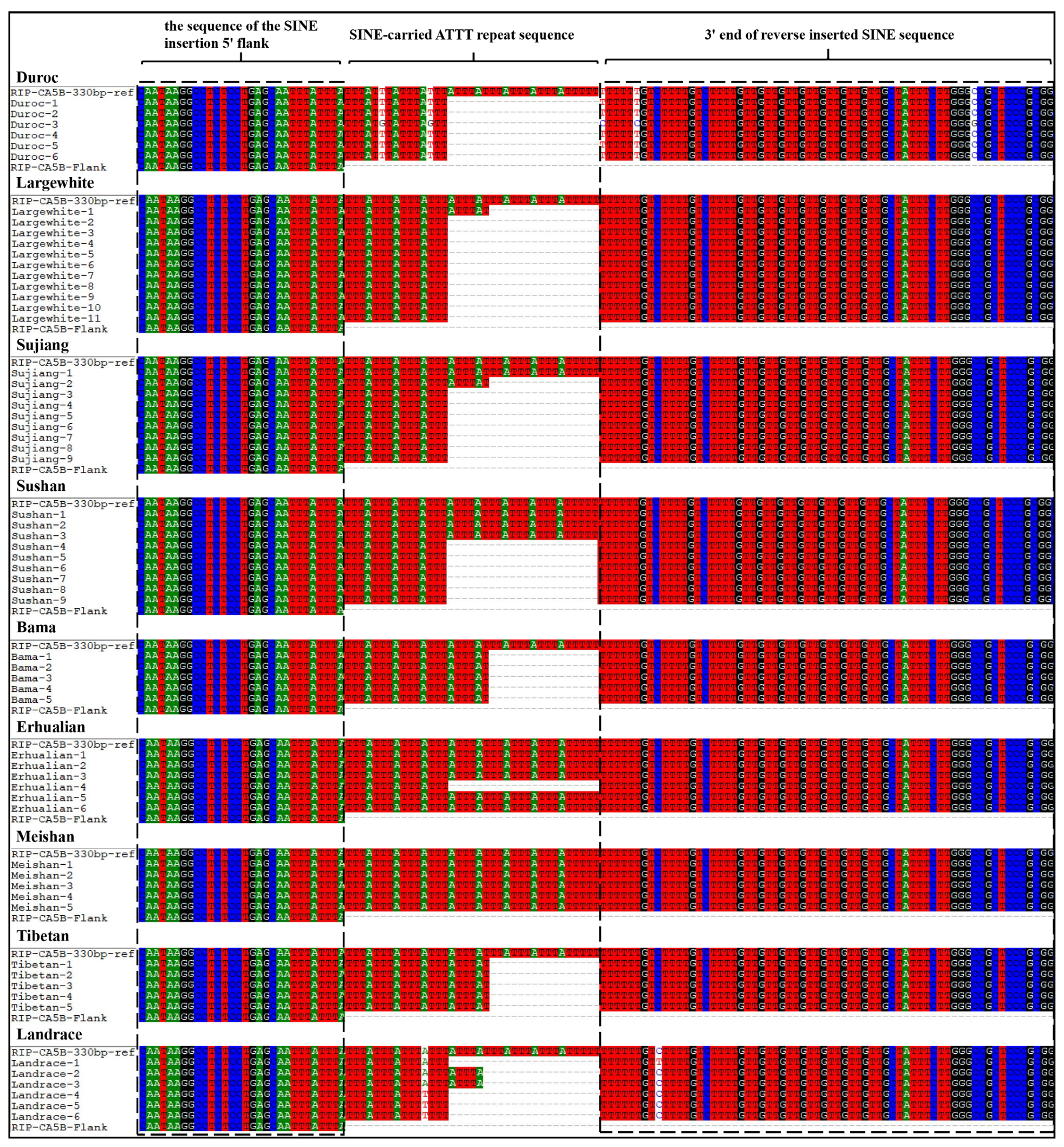
3.6. Length Variations of SINE-carried ATTT Repeat Sequence across Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef]
- Supuran, C.T.; Scozzafava, A. Carbonic anhydrase inhibitors and their therapeutic potential. Expert Opin. Ther. Pat. 2005, 10, 575–600. [Google Scholar] [CrossRef]
- Nishimori, I.; Minakuchi, T.; Onishi, S.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors. DNA cloning, characterization, and inhibition studies of the human secretory isoform VI, a new target for sulfonamide and sulfamate inhibitors. J. Med. Chem. 2007, 50, 381. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrase inhibitors as emerging drugs for the treatment of obesity. Expert Opin. Emerg. Drugs 2008, 13, 383. [Google Scholar] [CrossRef]
- Shah, G.N.; Hewett-Emmett, D.; Grubb, J.H. Mitochondrial carbonic anhydrase CA VB: Differences in tissue distribution and pattern of evolution from those of CA VA suggest distinct physiological roles. Proc. Natl. Acad. Sci. USA 2000, 97, 1677–1682. [Google Scholar] [CrossRef]
- Hazen, S.A.; Waheed, A.; Sly, W.S.; Lanoue, K.F.; Lynch, C.J. Differentiation-dependent expression of CA V and the role of carbonic anhydrase isozymes in pyruvate carboxylation in adipocytes. FASEB J. 1996, 10, 481–490. [Google Scholar] [CrossRef]
- Dodgson, S.J.; Forster, R.E. Inhibition of CA V decreases glucose synthesis from pyruvate. Arch. Biochem. Biophys. 1986, 251, 198–204. [Google Scholar] [CrossRef]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 2005, 247, 165–221. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. SINEs. Wiley Interdiscip. Rev. RNA 2011, 2, 772–786. [Google Scholar] [CrossRef]
- Chen, C.; Wang, W.; Wang, X.; Shen, D.; Wang, S.; Wang, Y.; Gao, B.; Wimmers, K.; Mao, J.; Li, K.; et al. Retrotransposons evolution and impact on lncRNA and protein coding genes in pigs. Mob. DNA 2019, 10, 19. [Google Scholar] [CrossRef]
- Burgess, D.J. Population genetics: Mobile elements across human populations. Nature Rev. Genet. 2013, 14, 370. [Google Scholar] [CrossRef]
- Witherspoon, D.J.; Zhang, Y.; Xing, J.; Watkins, W.S.; Ha, H.; Batzer, M.A.; Jorde, L.B. Mobile element scanning (ME-Scan) identifies thousands of novel Alu insertions in diverse human populations. Genome Res. 2013, 23, 1170–1181. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Symer, D.E. How do mammalian transposons induce genetic variation? A conceptual framework. BioEssays 2013, 35, 397–407. [Google Scholar] [CrossRef]
- Kratochwil, C.F.; Kautt, A.F.; Nater, A.; Härer, A.; Liang, Y.; Henning, F.; Meyer, A. An intronic transposon insertion associates with a trans-species color polymorphism in Midas cichlid fishes. Nat. Commun. 2022, 13, 296. [Google Scholar] [CrossRef]
- Li, J.; Davis, B.W.; Jern, P.; Dorshorst, B.J.; Siegel, P.B.; Andersson, L. Characterization of the endogenous retrovirus insertion in CYP19A1 associated with henny feathering in chicken. Mob. DNA 2019, 10, 38. [Google Scholar] [CrossRef]
- Vassetzky, N.S.; Borodulina, O.R.; Ustyantsev, I.G.; Sergei, A.; Kosushkin, S.A.; Kramerov, D.A. Analysis of SINE Families B2, Dip, and Ves with Special Reference to Polyadenylation Signals and Transcription Terminators. Int. J. Mol. Sci. 2021, 22, 9897. [Google Scholar] [CrossRef]
- Anna, D.S. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 2021, 22, 691–711. [Google Scholar]
- Wang, X.; Chi, C.; He, J.; Du, Z.; Zheng, Y.; D’Alessandro, E.; Chen, C.; Moawad, A.S.; Asare, E.; Song, C. SINE Insertion May Act as a Repressor to Affect the Expression of Pig LEPROT and Growth Traits. Genes 2022, 13, 1422. [Google Scholar] [CrossRef]
- Murphy, S.C.; Evans, J.M.; Tsai, K.L.; Clark, L.A. Length variations within the Merle retrotransposon of canine PMEL: Correlating genotype with phenotype. Mob. DNA 2018, 9, 26. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Wang, M.; Murani, E.; D’Alessandro, E.; Moawad, A.S.; Wang, X.; Wimmers, K.; Song, C. SINE Insertion in the Intron of Pig GHR May Decrease its Expression by Acting as a Repressor. Animals 2021, 1871, 1871. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, C.; Chen, W.; Wang, X.-Y.; Wang, W.; Gao, B.; Wimmers, K.; Mao, J.-D.; Song, C.-Y. Two new SINE insertion polymorphisms in pig Vertnin (VRTN) gene revealed by comparative genomic alignment. J. Integr. Agric. 2020, 19, 2514–2522. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2012, 41, D1063–D1069. [Google Scholar] [CrossRef]
- Singer, D.S.; Parent, L.J.; Ehrlich, R. Identification and DNA sequence of an interspersed repetitive DNA element in the genome of the miniature swine. Nucleic Acids Res. 1987, 15, 2780. [Google Scholar] [CrossRef]
- Prescott, D. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar]
- Chen, C.; D’alessandro, E.; Murani, E.; Zheng, Y.; Giosa, D.; Yang, N.; Wang, X.; Gao, B.; Li, K.; Wimmers, K.; et al. SINE jumping contributes to large-scale polymorphisms in the pig genomes. Mob. DNA 2021, 2021, 17. [Google Scholar] [CrossRef]
- Chan, S.; Shen, D.; Sang, Y.; Wang, S.; Wang, Y.; Chen, C.; Gao, B.; Song, C. Development of enhancer-trapping and -detection vectors mediated by the Tol2 transposon in zebrafish. PeerJ 2019, 7, e6862. [Google Scholar] [CrossRef]
- Kalla, S.E.; Moghadam, H.K.; Tomlinson, M.; Seebald, A.; Allen, J.J.; Whitney, J.; Choi, J.D.; Sutter, N.B. View ORCID ProfileNathan B. Sutter. Polymorphic SINEC_Cf Retrotransposons in the Genome of the Dog (Canis familiaris). bioRxiv 2020, 2020, 1–49. [Google Scholar]
- Rebollo, R.; Romanish, M.T.; Mager, D.L. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012, 46, 21–42. [Google Scholar] [CrossRef]
- Mita, P.; Boeke, J.D. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef]
- Shafee, T.; Lowe, R. Eukaryotic and prokaryotic gene structure. Wikijournal Med. 2017, 4, 1–5. [Google Scholar] [CrossRef]
- Xu, J.; Liu, T.; Li, D.; Zhang, Z.; Xia, Q.; Zhou, Z. BmSE, a SINE family with 3′ ends of (ATTT) repeats in domesticated silkworm (Bombyx mori). J. Genet. Genom. 2010, 37, 125–135. [Google Scholar] [CrossRef]
- Kosushkin, S.A.; Ustyantsev, I.G.; Borodulina, O.R.; Vassetzky, N.S.; Kramerov, D.A. Tail Wags Dog’s SINE: Retropositional Mechanisms of Can SINE Depend on Its A-Tail Structure. Biology 2022, 11, 1403. [Google Scholar] [CrossRef]
- Vedrine, S.M.; Vourc’h, P.; Tabagh, R.; Mignon, L.; Höfflin, S.; Cherpi-Antar, C.; Mbarek, O.; Paubel, A.; Moraine, C.; Raynaud, M.; et al. A functional tetranucleotide (AAAT) polymorphism in an Alu element in the NF1 gene is associated with mental retardation. Neurosci. Lett. 2011, 491, 118–121. [Google Scholar] [CrossRef]
- Marui, T.; Hashimoto, O.; Nanba, E.; Kato, C.; Tochigi, M.; Umekage, T.; Ishijima, M.; Kohda, K.; Kato, N.; Sasaki, T. Association between the neurofibromatosis-1 (NF1) locus and autism in the Japanese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004, 131, 43–47. [Google Scholar] [CrossRef]
- Plank, S.M.; Copeland-Yates, S.A.; Sossey-Alaoui, K.; Bell, J.M.; Schroer, R.J.; Skinner, C.; Michaeli, R.C. Lack of Association of the (AAAT)6 Allele of the GXAlu tetranucleotide Repeat in Intron 27b of the NF1 Gene with Autism. Am. J. Med. Genet. (Neuropsychiatr. Genet.) 2001, 105, 404–405. [Google Scholar] [CrossRef]


| Type | Breed | Province | Sample Size | Breeds Used in Creation of a Given Breed |
|---|---|---|---|---|
| Lean and hybrid | Sushan | Jiangsu | 32 | Meishan, Erhualian, and large white |
| Lean | Duroc | Anhui | 24 | / |
| Lean | Large White | Anhui | 506 | / |
| Lean | Landrace | Anhui | 24 | / |
| Medium and hybrid | Sujiang | Jiangsu | 24 | Jiangquhai, Fengjing, and Duroc |
| Fat | Meishan | Jiangsu | 24 | / |
| Fat | Fengjing | Jiangsu | 24 | / |
| Fat | Erhualian | Jiangsu | 24 | / |
| Miniature | Bama | Guangxi | 24 | / |
| Miniature | Banna | Yunnan | 24 | / |
| Miniature | Wuzhishan | Hainan | 24 | / |
| / | Wild boars | Anhui | 24 | / |
| Breed | Sample Size | Genotype Number | Genotype Frequency | Allele Frequency | Hardy–Weinberg Equilibrium Test/p Value | PIC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | +/+ | +/− | −/− | + | − | ||||
| Duroc | 24 | 10 | 6 | 8 | 0.42 | 0.25 | 0.33 | 0.54 | 0.46 | 0.015 | 0.373 |
| Landrace | 24 | 13 | 11 | 0 | 0.54 | 0.46 | 0.00 | 0.77 | 0.23 | 0.145 | 0.291 |
| Large White | 24 | 18 | 3 | 3 | 0.75 | 0.13 | 0.13 | 0.81 | 0.19 | 0.004 | 0.258 |
| Sujiang | 24 | 17 | 5 | 2 | 0.71 | 0.21 | 0.08 | 0.81 | 0.19 | 0.121 | 0.258 |
| Fengjing | 24 | 15 | 8 | 1 | 0.63 | 0.33 | 0.04 | 0.79 | 0.21 | 0.959 | 0.275 |
| Sushan | 32 | 5 | 8 | 19 | 0.16 | 0.25 | 0.59 | 0.28 | 0.72 | 0.031 | 0.323 |
| Meishan | 24 | 17 | 7 | 0 | 0.71 | 0.29 | 0.00 | 0.85 | 0.15 | 0.403 | 0.218 |
| Erhualian | 24 | 9 | 14 | 1 | 0.38 | 0.58 | 0.04 | 0.67 | 0.33 | 0.126 | 0.346 |
| Bama | 24 | 21 | 2 | 1 | 0.88 | 0.08 | 0.04 | 0.92 | 0.08 | 0.026 | 0.141 |
| Animal Numbers | Genotype | Genotype Frequency | Allele Frequency | Age at 100 kg Body Weight (Day) | Correcting Back Fat Thickness (cm) | |
|---|---|---|---|---|---|---|
| + | − | |||||
| 316 | +/+ | 0.66 | 0.80 | 0.20 | 163.25 ± 0.52 | 11.11 ± 0.14 a |
| 137 | +/− | 0.28 | 162.10 ± 0.52 | 11.00 ± 0.22 ab | ||
| 29 | −/− | 0.06 | 160.79 ± 1.07 | 10.15 ± 0.31 b | ||
| Breed-Individual Number | SV Length | Breed-Individual Number | SV Length | Breed-Individual Number | SV Length | Breed-Individual Number | SV Length |
|---|---|---|---|---|---|---|---|
| Duroc-1 | 308 bp | Sujiang-1 | 330 bp | Bama-1 | 314 bp | Tibetan-1 | 314 bp |
| Duroc-2 | 308 bp | Sujiang-2 | 314 bp | Bama-2 | 314 bp | Tibetan-2 | 314 bp |
| Duroc-3 | 308 bp | Sujiang-3 | 308 bp | Bama-3 | 314 bp | Tibetan-3 | 314 bp |
| Duroc-4 | 308 bp | Sujiang-4 | 308 bp | Bama-4 | 314 bp | Tibetan-4 | 314 bp |
| Duroc-5 | 308 bp | Sujiang-5 | 308 bp | Bama-5 | 314 bp | Tibetan-5 | 314 bp |
| Duroc-6 | 308 bp | Sujiang-6 | 308 bp | Erhualian-1 | 330 bp | Landrace-1 | 308 bp |
| Large white-1 | 314 bp | Sujiang-7 | 308 bp | Erhualian-2 | 330 bp | Landrace-2 | 313 bp |
| Large white-2 | 308 bp | Sujiang-8 | 308 bp | Erhualian-3 | 330 bp | Landrace-3 | 313 bp |
| Large white-3 | 308 bp | Sujiang-9 | 308 bp | Erhualian-4 | 308 bp | Landrace-4 | 308 bp |
| Large white-4 | 308 bp | Sushan-1 | 330 bp | Erhualian-5 | 330 bp | Landrace-5 | 308 bp |
| Large white-5 | 308 bp | Sushan-2 | 330 bp | Erhualian-6 | 330 bp | Landrace-6 | 308 bp |
| Large white-6 | 308 bp | Sushan-3 | 330 bp | Meishan-1 | 330 bp | ||
| Large white-7 | 308 bp | Sushan-4 | 308 bp | Meishan-2 | 330 bp | ||
| Large white-8 | 308 bp | Sushan-5 | 308 bp | Meishan-3 | 330 bp | ||
| Large white-9 | 308 bp | Sushan-6 | 308 bp | Meishan-4 | 330 bp | ||
| Large white-10 | 308 bp | Sushan-7 | 308 bp | Meishan-5 | 330 bp | ||
| Large white-11 | 308 bp | Sushan-8 | 308 bp | ||||
| Sushan-9 | 308 bp |
| SV Length | Number of Individuals | (ATTT)n * | total | % * | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duroc | Large white | Sujiang | Sushan | Bama | Erhualian | Meishan | Tibetan | Landrace | ||||
| 330 bp | 0 | 0 | 1 | 3 | 0 | 5 | 5 | 0 | 0 | (ATTT)9 | 14 | 23% |
| 314 bp | 0 | 1 | 1 | 0 | 5 | 0 | 0 | 5 | 0 | (ATTT)6 | 12 | 19% |
| 313 bp | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | (ATTT)6 | 2 | 3% |
| 308 bp | 6 | 10 | 7 | 6 | 0 | 1 | 0 | 0 | 4 | (ATTT)4 | 34 | 55% |
| total | 6 | 11 | 9 | 9 | 5 | 6 | 5 | 5 | 6 | / | 62 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Chen, C.; Wang, M.; Moawad, A.S.; Wang, X.; Song, C. SINE Insertion in the Pig Carbonic Anhydrase 5B (CA5B) Gene Is Associated with Changes in Gene Expression and Phenotypic Variation. Animals 2023, 13, 1942. https://doi.org/10.3390/ani13121942
Zheng Y, Chen C, Wang M, Moawad AS, Wang X, Song C. SINE Insertion in the Pig Carbonic Anhydrase 5B (CA5B) Gene Is Associated with Changes in Gene Expression and Phenotypic Variation. Animals. 2023; 13(12):1942. https://doi.org/10.3390/ani13121942
Chicago/Turabian StyleZheng, Yao, Cai Chen, Mengli Wang, Ali Shoaib Moawad, Xiaoyan Wang, and Chengyi Song. 2023. "SINE Insertion in the Pig Carbonic Anhydrase 5B (CA5B) Gene Is Associated with Changes in Gene Expression and Phenotypic Variation" Animals 13, no. 12: 1942. https://doi.org/10.3390/ani13121942
APA StyleZheng, Y., Chen, C., Wang, M., Moawad, A. S., Wang, X., & Song, C. (2023). SINE Insertion in the Pig Carbonic Anhydrase 5B (CA5B) Gene Is Associated with Changes in Gene Expression and Phenotypic Variation. Animals, 13(12), 1942. https://doi.org/10.3390/ani13121942








