Haplotypes of the tRNAleu-COII mtDNA Region in Russian Apis mellifera Populations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
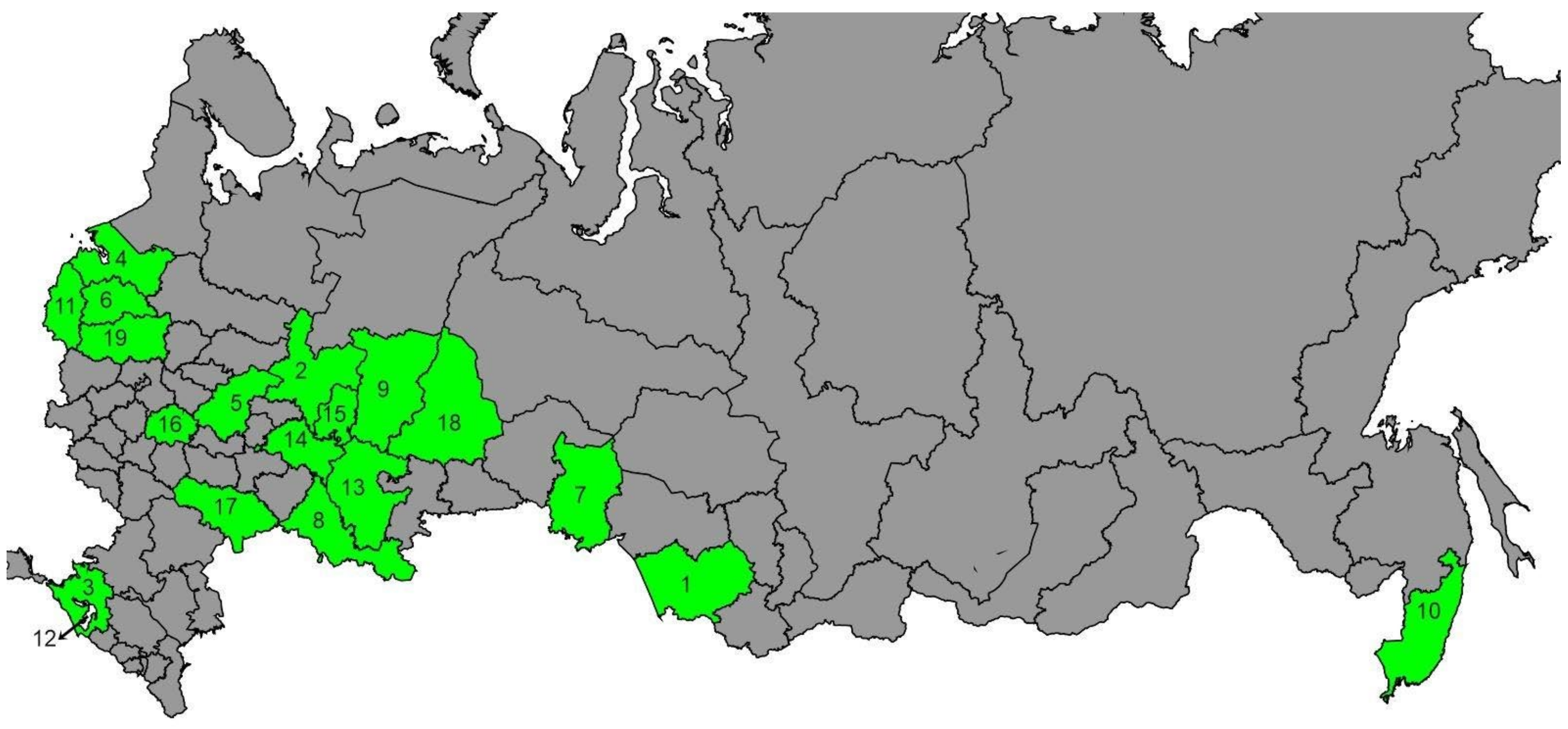
2.1. Sampling and DNA Extraction
2.2. tRNAleu-COII Locus Analysis
3. Results
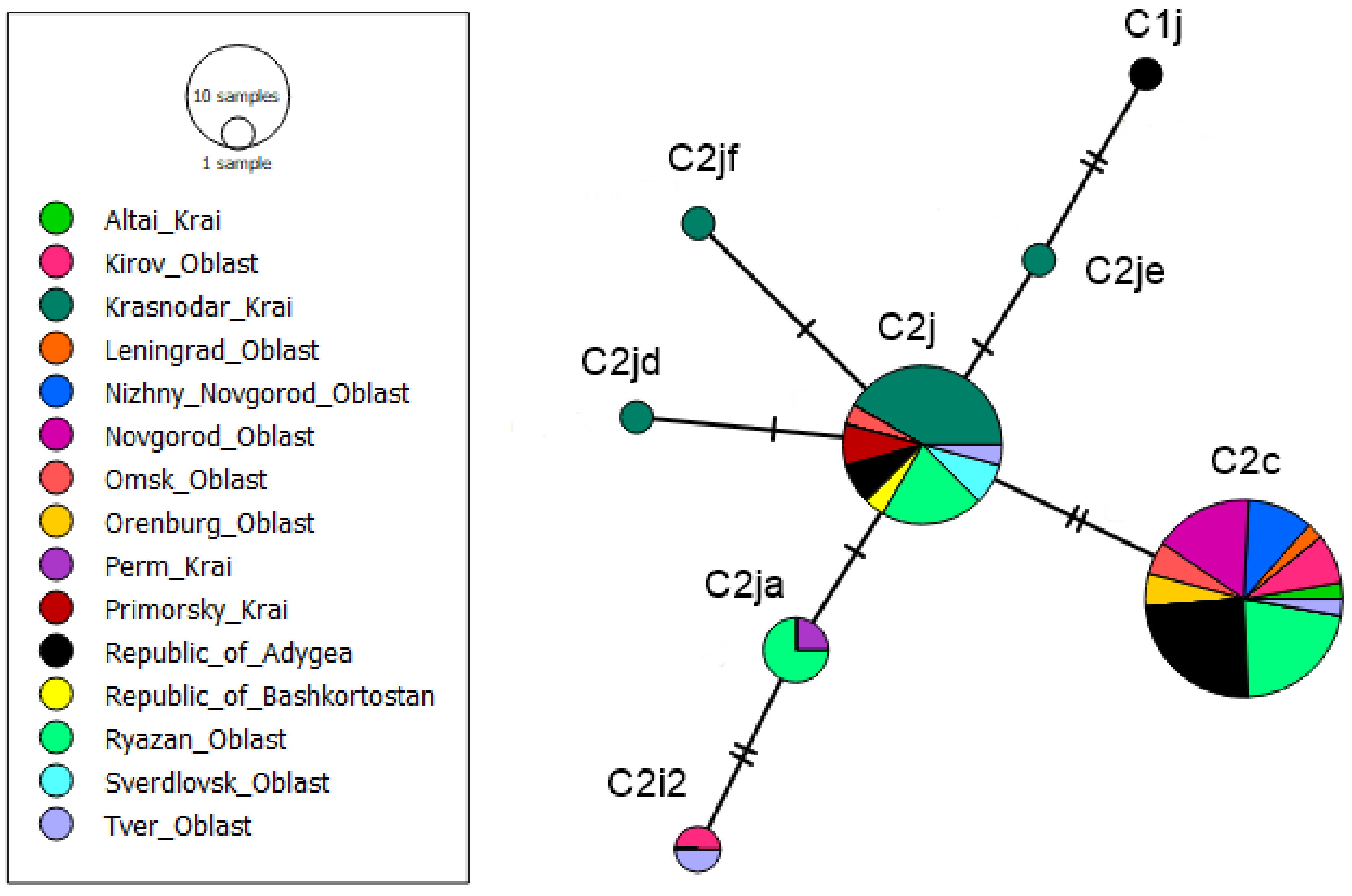
3.1. tRNAleu-COII Haplotypes from Evolutionary Lineage C
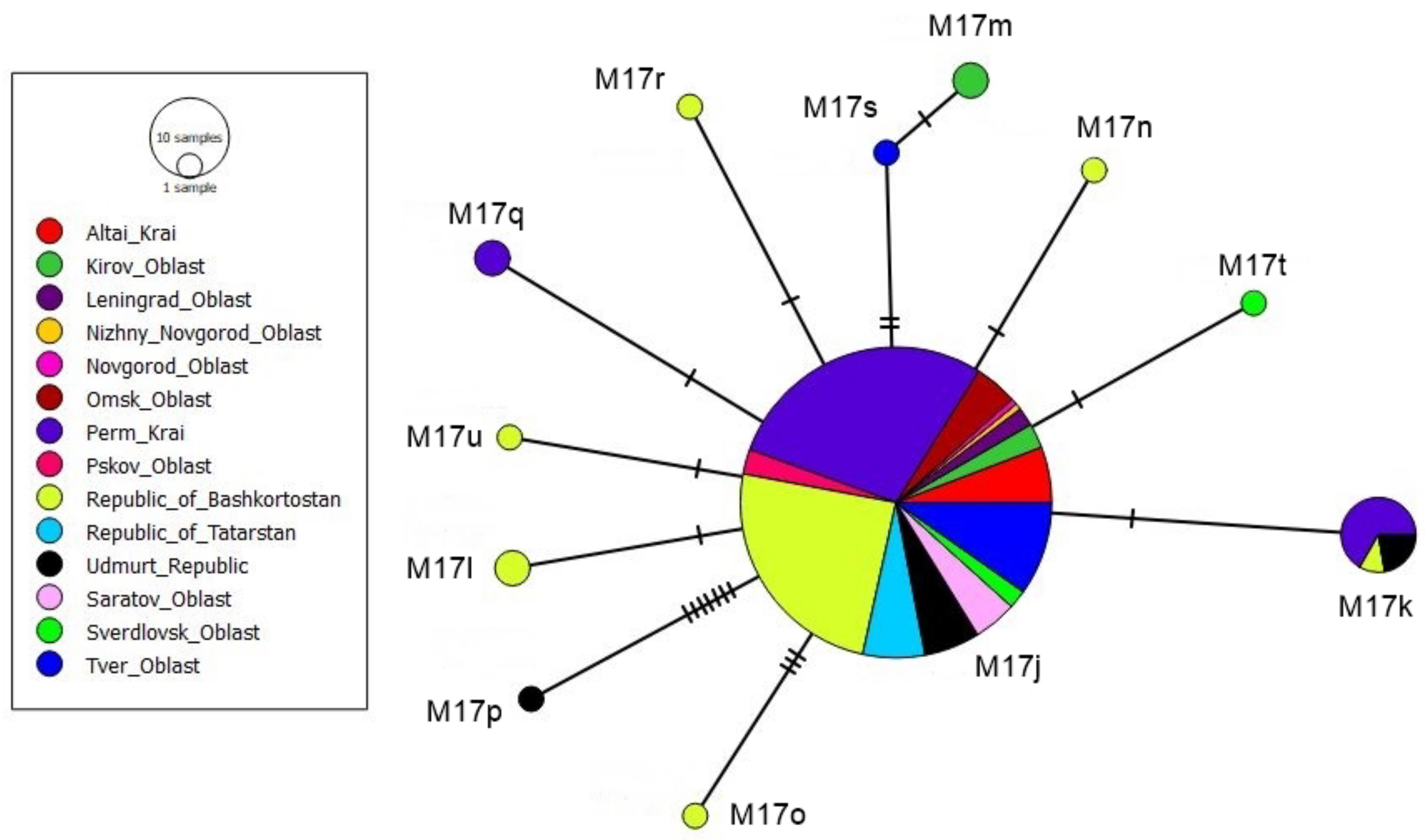
3.2. tRNAleu-COII Haplotypes from Evolutionary Lineage M
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jensen, A.B.; Palmer, K.A.; Boomsma, J.J.; Pedersen, B.V. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005, 14, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Oleksa, A.; Chybicki, I.; Tofilski, A.; Burczyk, J. Nuclear and mitochondrial patterns of introgression into native dark bees (Apis mellifera mellifera) in Poland. J. Apic. Res. 2011, 50, 116–129. [Google Scholar] [CrossRef] [Green Version]
- Nolte, A.W.; Tautz, D. Understanding the onset of hybrid speciation. Trends Genet. 2009, 26, 54–58. [Google Scholar] [CrossRef]
- Rhymer, J.M.; Simberloff, D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Evol. Syst. 1996, 27, 83–109. [Google Scholar] [CrossRef]
- Buchler, R.; Costa, C.; Hatjina, F.; Andonov, S.; Meixner, M.D.; Conte, Y.L.; Uzunov, A.; Berg, S.; Bienkowska, M.; Bouga, M.; et al. The influence of genetic origin and its interaction with environmental effects on the survival of Apis mellifera L. colonies in Europe. J. Apic. Res. 2014, 53, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Munoz, I.; Henriques, D.; Jara, L.; Johnston, J.S.; Chavez-Galarza, J.; De La Rua, P.; Pinto, M.A. SNPs selected by information content outperform randomly selected microsatellite loci for delineating genetic identification and introgression in the endangered dark European honeybee (Apis mellifera mellifera). Mol. Ecol. Resour. 2017, 17, 783–795. [Google Scholar] [CrossRef] [Green Version]
- Alpatov, V.V. Bee races and red clover pollination. Bee World 1948, 29, 61–63. [Google Scholar] [CrossRef]
- Ruttner, F. Biogeography and Taxonomy of Honeybees; Springer: Berlin, Germany, 1988; p. 291. [Google Scholar] [CrossRef]
- Nikolenko, A.G.; Poskryakov, A.V. Polymorphism of Locus COI-COII of Mitochondrial DNA in the Honeybee Apis mellifera L. from the Southern Ural Region. Russ. J. Genet. 2002, 38, 364–368. [Google Scholar] [CrossRef]
- Ilyasov, R.A.; Petukhov, A.V.; Poskryakov, A.V.; Nikolenko, A.G. Local honeybee (Apis mellifera mellifera L.) populations in the Urals. Russ. J. Genet. 2007, 43, 709–711. [Google Scholar] [CrossRef]
- Pedersen, B.V. On the phylogenetic position of the Danish strain of the black honeybee (the Læsø bee), Apis mellifera mellifera L.(Hymenoptera: Apidae) inferred from mitochondrial DNA sequences. Insect Syst. Evol. 1996, 27, 241–250. [Google Scholar] [CrossRef]
- De la Rua, P.; Jaffe, R.; Dall’Olio, R.; Munoz, I.; Serrano, J. Biodiversity, conservation and current threats to European honeybees. Apidologie 2009, 40, 263–284. [Google Scholar] [CrossRef] [Green Version]
- Kryger, P. New threats for honeybee conservation on the island of Læso. In Proceedings of the 5th European Conference of Apidology, Halle an der Saale, Germany, 3–7 September 2012; Volume 5, p. 134. [Google Scholar]
- Ruottinen, L.; Berg, P.; Kantanen, J.; Kristensen, T.N.; Præbel, A. Status and Conservation of the Nordic Brown Bee: Final report. NordGen 2014, 42, 5–35. [Google Scholar] [CrossRef]
- Arias, M.C.; Sheppard, W.S. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol. Phylogenetics Evol. 1996, 5, 557–566. [Google Scholar] [CrossRef]
- Cridland, J.M.; Tsutsui, N.D.; Ramírez, S.R. The complex demographic history and evolutionary origin of the western honey bee, Apis mellifera. Genome Biol. Evol. 2017, 9, 457–472. [Google Scholar] [CrossRef] [Green Version]
- Momeni, J.; Parejo, M.; Nielsen, R.O.; Langa, J.; Montes, I.; Papoutsis, L.; Farajzadeh, L.; Bendixen, C.; Căuia, E.; Charrière, J.-D.; et al. Authoritative subspecies diagnosis tool for European honey bees based on ancestry informative SNPs. BMC Genom. 2021, 22, 101. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.W.; Behura, S.K.; Berlocher, S.H.; Clark, A.G.; Johnston, J.S.; Sheppard, W.S.; Smith, D.R.; Suarez, A.V.; Weaver, D.; Tsutsui, N.D. Thrice out of Africa: Ancient and recent expansions of the honey bee, Apis mellifera. Science 2006, 314, 642–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallberg, A.; Han, F.; Wellhagen, G.; Dahle, B.; Kawata, M.; Haddad, N.; Simões, Z.L.P.; Allsopp, M.H.; Kandemir, I.; De la Rúa, P. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 2014, 46, 1081–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garnery, L.; Solignac, M.; Celebrano, G.; Cornuet, J.-M. A simple test using restricted PCR-amplified mitochondrial DNA to study the genetic structure of Apis mellifera L. Experientia 1993, 49, 1016–1021. [Google Scholar] [CrossRef]
- Garnery, L.; Cornuet, J.M.; Solignac, M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992, 1, 145–154. [Google Scholar] [CrossRef]
- Franck, P.; Garnery, L.; Loiseau, A.; Oldroyd, B.P.; Hepburn, H.R.; Solignac, M.; Cornuet, J.M. Genetic diversity of the honeybee in Africa: Microsatellite and mitochondrial data. Heredity 2001, 86, 420–430. [Google Scholar] [CrossRef] [Green Version]
- Shaibi, T.; Muñoz, I.; Dall′ Olio, R.; Lodesani, M.; De la Rúa, P.; Moritz, R.F.A. Apis mellifera evolutionary lineages in Northern Africa: Libya, where orient meets occident. Insect. Soc. 2009, 56, 293–300. [Google Scholar] [CrossRef]
- Meixner, M.D.; Pinto, M.A.; Bouga, M.; Kryger, P.; Ivanova, E.; Fuchs, S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Oleksa, A.; Kusza, S.; Tofilski, A. Mitochondrial DNA Suggests the Introduction of Honeybees of African Ancestry to East-Central Europe. Insects 2021, 12, 410. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Garnery, L. Mitochondrial DNA variability in honeybees and its phylogeographic implications. Apidologie 1991, 22, 627–642. [Google Scholar] [CrossRef] [Green Version]
- Rortais, A.; Arnold, G.; Alburaki, M.; Legout, H.; Garnery, L. Review of the DraI COI-COII test for the conservation of the black honeybee (Apis mellifera mellifera). Conserv. Genet. Resour. 2011, 3, 383–391. [Google Scholar] [CrossRef]
- Hassett, J.; Browne, K.A.; McCormack, G.P.; Moore, E.; Society, N.I.H.B.; Soland, G.; Geary, M. A significant pure population of the dark European honey bee (Apis mellifera mellifera) remains in Ireland. J. Apic. Res. 2018, 57, 337–350. [Google Scholar] [CrossRef] [Green Version]
- Chávez-Galarza, J.; López-Montañez, R.; Jiménez, A.; Ferro-Mauricio, R.; Oré, J.; Medina, S.; Rea, R.; Vásquez, H. Mitochondrial DNA Variation in Peruvian Honey Bee (Apis mellifera L.) Populations Using the tRNAleu-cox2 Intergenic Region. Insects 2021, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Collet, T.; Ferreira, K.; Arias, M.; Soares, A.E.E.; Del Lama, M.A. Genetic structure of Africanized honeybee populations (Apis mellifera L.) from Brazil and Uruguay viewed through mitochondrial DNA COI–COII patterns. Heredity 2006, 97, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alburaki, M.; Madella, S.; Lopez, J.; Bouga, M.; Chen, Y.; vanEngelsdorp, D. Honey bee populations of the USA display restrictions in their mtDNA haplotype diversity. Front. Genet. 2023, 13, 1092121. [Google Scholar] [CrossRef] [PubMed]
- Kaskinova, M.D.; Gaifullina, L.R.; Saltykova, E.S.; Poskryakov, A.V.; Nikolenko, A.G. Dynamics of the Genetic Structure of Apis mellifera Populations in the Southern Urals. Russ. J. Genet. 2022, 58, 36–41. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Tanasković, M.; Erić, P.; Patenković, A.; Erić, K.; Mihajlović, M.; Tanasić, V.; Stanisavljević, L.; Davidović, S. MtDNA Analysis Indicates Human-Induced Temporal Changes of Serbian Honey Bees Diversity. Insects 2021, 12, 767. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Galarza, J.; Garnery, L.; Henriques, D.; Neves, C.J.; Loucif-Ayad, W.; Jonhston, J.S.; Pinto, M.A. Mitochondrial DNA variation of Apis mellifera iberiensis: Further insights from a large scale study using sequence data of the tRNAleu-cox2 intergenic region. Apidologie 2017, 48, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Galarza, J.; Henriques, D.; Johnston, J.S.; Carneiro, M.; Rufino, J.; Patton, J.C.; Pinto, M.A. Revisiting the Iberian honey bee (Apis mellifera iberiensis) contact zone: Maternal and genome-wide nuclear variations provide support for secondary contact from historical refugia. Mol. Ecol. 2015, 24, 2973–2992. [Google Scholar] [CrossRef] [PubMed]
- Krivcov, N.I.; Sokol’skij, S.S.; Lyubimov, E.M. Serye Gornye Kavkazskie Pchyoly; Nauchnoe Izd: Sochi, Russia, 2009; 192p. (In Russian) [Google Scholar]
- Soland-Reckeweg, G.; Heckel, G.; Neumann, P.; Fluri, P.; Excoffier, L. Gene flow in admixed populations and implications for the conservation of the Western honeybee, Apis mellifera. J. Insect Conserv. 2009, 13, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Parejo, M.; Wragg, D.; Gauthier, L.; Vignal, A.; Neumann, P.; Neuditschko, M. Using Whole-Genome Sequence Information to Foster Conservation Efforts for the European Dark Honey Bee, Apis mellifera mellifera. Front. Ecol. Evol. 2016, 4, 583. [Google Scholar] [CrossRef] [Green Version]
- Bouga, M.; Alaux, C.; Bienkowska, M.; Büchler, R.; Carreck, N.L.; Cauia, E.; Chlebo, R.; Dahle, B.; Dall’Olio, R.; De la Rúa, P.; et al. A review of methods for discrimination of honey bee populations as applied to European beekeeping. J. Apic. Res. 2011, 50, 51–84. [Google Scholar] [CrossRef] [Green Version]
- Strange, J.P.; Garnery, L.; Sheppard, W.S. Persistence of the Landes ecotype of Apis mellifera mellifera in southwest France: Confirmation of a locally adaptive annual brood cycle trait. Apidologie 2007, 38, 259–267. [Google Scholar] [CrossRef] [Green Version]
- Thompson, C. The Health and Status of the Feral Honeybee (Apis mellifera sp.) and Apis mellifera mellifera Population of the UK. Ph.D. Thesis, University of Leeds, Leeds, UK, 2010; p. 173. [Google Scholar]
- Soland-Reckeweg, G. Genetic Differentiation and Hybridization in the Honeybee (Apis mellifera L.) in Switzerland. Ph.D. Thesis, University of Bern, Bern, Switzerland, 2006; 72p. [Google Scholar]
- Meixner, M.D.; Worobik, M.; Wilde, J.; Fuchs, S.; Koeniger, N. Apis mellifera mellifera in eastern Europe—Morphometric variation and determination of its range limits. Apidologie 2007, 38, 191–197. [Google Scholar] [CrossRef]
- Rortais, A.; Arnold, G.; Garnery, L. Utilisation des marqueurs mole’culaires pour la mise en place de conservatoires d’abeilles noires (Apis mellifera mellifera). Me’moires SEF 2009, 8, 115–120. [Google Scholar]

| DraI Restriction Sites | Haplotypes | Haplogroup |
|---|---|---|
| 47/40/64/420 | C2, C2c, C2l, C2j, C2ja, C2i2, C2jf, C2jd, C2je | C2 |
| 47/41/64/420 | C1, C1j | C1 |
| 47/40/63/420 | C3 | C3 |
| 47/39/64/420 | C4a | C4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaskinova, M.D.; Gaifullina, L.R.; Saltykova, E.S. Haplotypes of the tRNAleu-COII mtDNA Region in Russian Apis mellifera Populations. Animals 2023, 13, 2394. https://doi.org/10.3390/ani13142394
Kaskinova MD, Gaifullina LR, Saltykova ES. Haplotypes of the tRNAleu-COII mtDNA Region in Russian Apis mellifera Populations. Animals. 2023; 13(14):2394. https://doi.org/10.3390/ani13142394
Chicago/Turabian StyleKaskinova, Milyausha D., Luisa R. Gaifullina, and Elena S. Saltykova. 2023. "Haplotypes of the tRNAleu-COII mtDNA Region in Russian Apis mellifera Populations" Animals 13, no. 14: 2394. https://doi.org/10.3390/ani13142394
APA StyleKaskinova, M. D., Gaifullina, L. R., & Saltykova, E. S. (2023). Haplotypes of the tRNAleu-COII mtDNA Region in Russian Apis mellifera Populations. Animals, 13(14), 2394. https://doi.org/10.3390/ani13142394





