Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
2.2. PCR
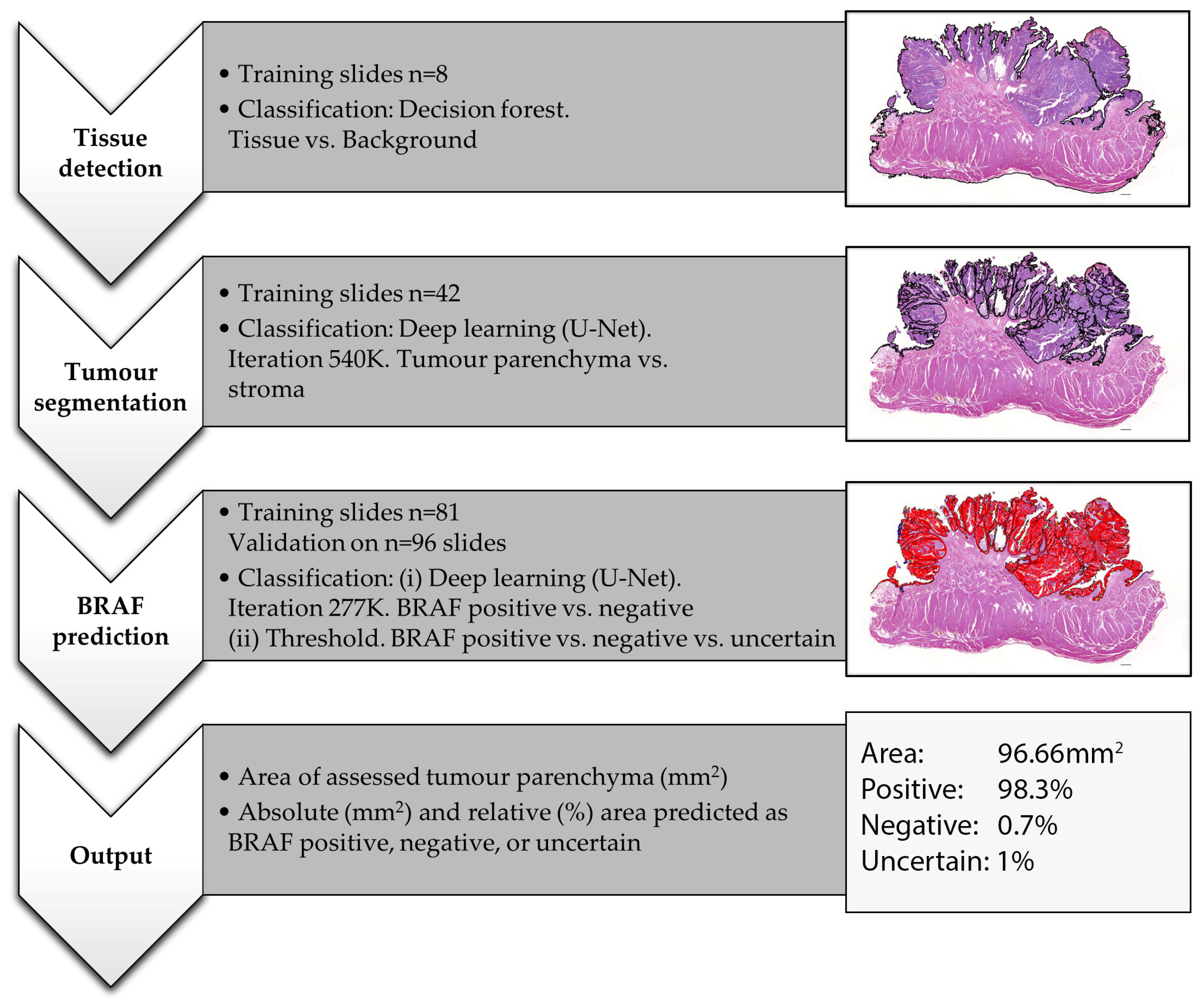
2.3. AI Histology
- (i)
- Tissue detection.
- (ii)
- Tissue and tumour segmentation.
- (iii)
- BRAF mutation prediction
- (1)
- Threshold 0.6: positive (≥0.6 probability), negative (≥0.6 probability) or uncertain (<0.6 positive and <0.6 negative);
- (2)
- Threshold 0.7: positive (≥0.7 probability), negative (≥0.7 probability) or uncertain (<0.7 positive and <0.7 negative);
- (3)
- Threshold 0.5: positive (≥0.5 probability) or negative (≥0.5 probability).
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jung, H.; Bae, K.; Lee, J.Y.; Kim, J.H.; Han, H.J.; Yoon, H.Y.; Yoon, K.A. Establishment of canine transitional cell carcinoma cell lines harboring BRAF V595E mutation as a therapeutic target. Int. J. Mol. Sci. 2021, 22, 9151. [Google Scholar] [CrossRef]
- De Brot, S.; Robinson, B.D.; Scase, T.; Grau-Roma, L.; Wilkinson, E.; Boorjian, S.A.; Gardner, D.; Mongan, N.P. The dog as an animal model for bladder and urethral urothelial carcinoma: Comparative epidemiology and histology. Oncol. Lett. 2018, 16, 1641–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grassinger, J.M.; Merz, S.; Aupperle-Lellbach, H.; Erhard, H.; Klopfleisch, R. Correlation of BRAF variant V595E, breed, histological grade and cyclooxygenase-2 expression in canine transitional cell carcinomas. Vet. Sci. 2019, 6, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, D.W.; Dhawan, D.; Ramos-Vara, J.A.; Ratliff, T.L.; Cresswell, G.M.; Utturkar, S.; Sommer, B.C.; Fulkerson, C.M.; Hahn, N.M. Naturally-Occurring Invasive Urothelial Carcinoma in Dogs, a Unique Model to Drive Advances in Managing Muscle Invasive Bladder Cancer in Humans. Front. Oncol. 2020, 9, 1493. [Google Scholar] [CrossRef]
- De Brot, S.; Grau-Roma, L.; Stirling-Stainsby, C.; Dettwiler, M.; Guscetti, F.; Meier, D.; Scase, T.; Robinson, B.D.; Gardner, D.; Mongan, N.P. A Fibromyxoid Stromal Response is Associated with Muscle Invasion in Canine Urothelial Carcinoma. J. Comp. Pathol. 2019, 169, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, W. Tumors in Domestic Animals, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Norris, A.M.; Laing, E.J.; Valli, V.E.O.; Withrow, S.J.; Macy, D.W.; Ogilvie, G.K.; Tomlinson, J.; McCaw, D.; Pidgeon, G.; Jacobs, R.M. Canine Bladder and Urethral Tumors: A Retrospective Study of 115 Cases (1980–1985). J. Vet. Intern. Med. 1992, 6, 145–153. [Google Scholar] [CrossRef]
- Fulkerson, C.M.; Knapp, D.W. Management of transitional cell carcinoma of the urinary bladder in dogs: A review. Vet. J. 2015, 205, 217–225. [Google Scholar] [CrossRef]
- Fulkerson, C.M.; Dhawan, D.; Ratliff, T.L.; Hahn, N.M.; Knapp, D.W. Naturally Occurring Canine Invasive Urinary Bladder Cancer: A Complementary Animal Model to Improve the Success Rate in Human Clinical Trials of New Cancer Drugs. Int. J. Genom. 2017, 2017, 6589529. [Google Scholar] [CrossRef]
- Mochizuki, H.; Kennedy, K.; Shapiro, S.G.; Breen, M.B. BRAF mutations in canine cancers. PLoS ONE 2015, 10, e0129534. [Google Scholar] [CrossRef] [Green Version]
- Ostrander, E.; Decker, B.; Parker, H.G.; Dhawan, D.; Kwon, E.M.; Karlins, E.; Davis, B.; Ramos-vara, J.A.; Bonney, P.L.; McNiel, E.A.; et al. Homologous Mutation to Human BRAF V600E is Common in Naturally Occurring Canine Bladder Cancer—Evidence for a Relevant Model System and Urine-based Diagnostic Test. Mol. Cancer Res. 2012, 17, 1310–1314. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, H.; Breen, M. Comparative aspects of BRAF mutations in canine cancers. Vet. Sci. 2015, 2, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, D.; Yashashwi, K.; Kumar, N.; Rane, S.; Gann, P.H.; Sethi, A. Weakly supervised learning on unannotated H&E-stained slides predicts BRAF mutation in thyroid cancer with high accuracy. J. Pathol. 2021, 255, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Alzahrani, A.S.; Carson, K.A.; Shong, Y.K.; Kim, T.Y.; Viola, D.; Robenshtok, E.; Fagin, J.A.; Puxeddu, E.; Fugazzola, L.; et al. Association Between BRAF V600E Mutation and Recurrence of Papillary Thyroid Cancer. J. Clin. Oncol. 2015, 33, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Lee, E.; Koo, J.S.; Yoon, J.H.; Nam, K.H.; Lee, J.; Jo, Y.S.; Moon, H.J.; Park, V.Y.; Kwak, J.Y. Artificial intelligence to predict the BRAFV600E mutation in patients with thyroid cancer. PLoS ONE 2020, 15, e0242806. [Google Scholar] [CrossRef]
- Ito, T.; Tanaka, Y.; Murata, M.; Kaku-Ito, Y.; Furue, K.; Furue, M. BRAF Heterogeneity in Melanoma. Curr. Treat. Options Oncol. 2021, 22, 20. [Google Scholar] [CrossRef]
- Peralta, S.; Webb, S.M.; Katt, W.P.; Grenier, J.K.; Duhamel, G.E. Highly recurrent BRAF p.V595E mutation in canine papillary oral squamous cell carcinoma. Vet. Comp. Oncol. 2023, 21, 138–144. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Bindal, N.; Bamford, S.; Ward, S.; Cole, C.G.; Jia, M.; Kok, C.; Boutselakis, H.; De, T.; et al. High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer. Curr. Protoc. Hum. Genet. 2016, 91. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Katherine, A.; Akbani, R.; et al. HHS Public Access bladder cancer. Cell 2018, 171, 540–556. [Google Scholar] [CrossRef]
- Iyer, G.; Al-Ahmadie, H.; Schultz, N.; Hanrahan, A.J.; Ostrovnaya, I.; Balar, A.V.; Kim, P.H.; Lin, O.; Weinhold, N.; Sander, C.; et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J. Clin. Oncol. 2013, 31, 3133–3140. [Google Scholar] [CrossRef]
- Boulalas, I.; Zaravinos, A.; Delakas, D.; Spandidos, D.A. Mutational analysis of the BRAF gene in transitional cell carcinoma of the bladder. Int. J. Biol. Markers 2009, 24, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Longo, T.; McGinley, K.F.; Freedman, J.A.; Etienne, W.; Wu, Y.; Sibley, A.; Owzar, K.; Gresham, J.; Moy, C.; Szabo, S.; et al. Targeted Exome Sequencing of the Cancer Genome in Patients with Very High-risk Bladder Cancer. Eur. Urol. 2016, 70, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Wiley, C.A.; Droste, E.L.; Robertson, J.; Inman, B.A.; Breen, M. Whole exome sequencing analysis of canine urothelial carcinomas without BRAF V595E mutation: Short in-frame deletions in BRAF and MAP2K1 suggest alternative mechanisms for MAPK pathway disruption. PLoS Genet. 2023, 19, e1010575. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, H.; Shapiro, S.G.; Breen, M. Detection of BRAF mutation in urine DNA as a molecular diagnostic for canine urothelial and prostatic carcinoma. PLoS ONE 2015, 10, e0144170. [Google Scholar] [CrossRef]
- Rasteiro, A.M.; Sá E Lemos, E.; Oliveira, P.A.; Gil da Costa, R.M. Molecular Markers in Urinary Bladder Cancer: Applications for Diagnosis, Prognosis and Therapy. Vet. Sci. 2022, 9, 107. [Google Scholar] [CrossRef]
- Davis, A.S.; Chang, M.Y.; Brune, J.E.; Hallstrand, T.S.; Johnson, B.; Lindhartsen, S.; Hewitt, S.M.; Frevert, C.W. The Use of Quantitative Digital Pathology to Measure Proteoglycan and Glycosaminoglycan Expression and Accumulation in Healthy and Diseased Tissues. J. Histochem. Cytochem. 2021, 69, 137–155. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef] [Green Version]
- Qu, H.; Zhou, M.; Yan, Z.; Wang, H.; Rustgi, V.K.; Zhang, S.; Gevaert, O.; Metaxas, D.N. Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. npj Precis. Oncol. 2021, 5, 87. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. npj Precis. Oncol. 2020, 4, 14. [Google Scholar] [CrossRef]
- Zuraw, A.; Aeffner, F. Whole-slide imaging, tissue image analysis, and artificial intelligence in veterinary pathology: An updated introduction and review. Vet. Pathol. 2022, 59, 6–25. [Google Scholar] [CrossRef]
- Hespel, A.M.; Zhang, Y.; Basran, P.S. Artificial intelligence 101 for veterinary diagnostic imaging. Vet. Radiol. Ultrasound 2022, 63, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.; O’Leary, M.; Yuan, L. Artificial intelligence in educational assessment: ‘Breakthrough? Or buncombe and ballyhoo?’. J. Comput. Assist. Learn. 2021, 37, 1207–1216. [Google Scholar] [CrossRef]
- Lustgarten, J.L.; Zehnder, A.; Shipman, W.; Gancher, E.; Webb, T.L. Veterinary informatics: Forging the future between veterinary medicine, human medicine, and One Health initiatives-a joint paper by the Association for Veterinary Informatics (AVI) and the CTSA One Health Alliance (COHA). JAMIA Open 2020, 3, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gedon, J.; Kehl, A.; Aupperle-Lellbach, H.; von Bomhard, W.; Schmidt, J.M. BRAF mutation status and its prognostic significance in 79 canine urothelial carcinomas: A retrospective study (2006–2019). Vet. Comp. Oncol. 2022, 20, 449–457. [Google Scholar] [CrossRef]
- Kiupel, M.; Camus, M. Diagnosis and Prognosis of Canine Cutaneous Mast Cell Tumors. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 819–836. [Google Scholar] [CrossRef]
- Tamlin, V.S.; Bottema, C.D.K.; Peaston, A.E. Comparative aspects of mast cell neoplasia in animals and the role of KIT in prognosis and treatment. Vet. Med. Sci. 2020, 6, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Bertram, C.A.; Aubreville, M.; Donovan, T.A.; Bartel, A.; Wilm, F.; Marzahl, C.; Assenmacher, C.A.; Becker, K.; Bennett, M.; Corner, S.; et al. Computer-assisted mitotic count using a deep learning-based algorithm improves interobserver reproducibility and accuracy. Vet. Pathol. 2022, 59, 211–226. [Google Scholar] [CrossRef]
- Shmatko, A.; Ghaffari Laleh, N.; Gerstung, M.; Kather, J.N. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef]
- Figueroa-Silva, O.; Pastur Romay, L.A.; Viruez Roca, R.D.; Rojas, M.D.S.A.Y.; Suárez-Peñaranda, J.M. Machine Learning Techniques in Predicting BRAF Mutation Status in Cutaneous Melanoma From Clinical and Histopathologic Features. Appl. Immunohistochem. Mol. Morphol. 2022, 30, 674–680. [Google Scholar] [CrossRef]
- Krebs, F.S.; Britschgi, C.; Pradervand, S.; Achermann, R.; Tsantoulis, P.; Haefliger, S.; Wicki, A.; Michielin, O.; Zoete, V. Structure-based prediction of BRAF mutation classes using machine-learning approaches. Sci. Rep. 2022, 12, 12528. [Google Scholar] [CrossRef]
- Kim, R.H.; Nomikou, S.; Coudray, N.; Jour, G.; Dawood, Z.; Hong, R.; Esteva, E.; Sakellaropoulos, T.; Donnelly, D.; Moran, U.; et al. Deep Learning and Pathomics Analyses Reveal Cell Nuclei as Important Features for Mutation Prediction of BRAF-Mutated Melanomas. J. Investig. Dermatol. 2022, 142, 1650–1658.e6. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, O.L.; Quirke, P.; West, N.P.; James, J.A.; Loughrey, M.B.; Grabsch, H.I.; Salto-Tellez, M.; Alwers, E.; Cifci, D.; Ghaffari Laleh, N.; et al. Swarm learning for decentralized artificial intelligence in cancer histopathology. Nat. Med. 2022, 28, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, M.; Browning, L.; Sirinukunwattana, K.; Malacrino, S.; Khalid Alham, N.; Colling, R.; Cui, Y.; Rakha, E.; Hamdy, F.C.; Verrill, C.; et al. Automated quality assessment of large digitised histology cohorts by artificial intelligence. Sci. Rep. 2022, 12, 5002. [Google Scholar] [CrossRef]
- Salvi, M.; Acharya, U.R.; Molinari, F.; Meiburger, K.M. The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis. Comput. Biol. Med. 2021, 128, 104129. [Google Scholar] [CrossRef] [PubMed]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Terada, Y.; Takahashi, T.; Hayakawa, T.; Ono, A.; Kawata, T.; Isaka, M.; Muramatsu, K.; Tone, K.; Kodama, H.; Imai, T.; et al. Artificial Intelligence-Powered Prediction of ALK Gene Rearrangement in Patients with Non–Small-Cell Lung Cancer. JCO Clin. Cancer Inform. 2022, 6, e2200070. [Google Scholar] [CrossRef]
- Nero, C.; Boldrini, L.; Lenkowicz, J.; Giudice, M.T.; Piermattei, A.; Inzani, F.; Pasciuto, T.; Minucci, A.; Fagotti, A.; Zannoni, G.; et al. Deep-Learning to Predict BRCA Mutation and Survival from Digital H&E Slides of Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 11326. [Google Scholar] [CrossRef]
- Yamashita, R.; Long, J.; Longacre, T.; Peng, L.; Berry, G.; Martin, B.; Higgins, J.; Rubin, D.L.; Shen, J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: A diagnostic study. Lancet Oncol. 2021, 22, 132–141. [Google Scholar] [CrossRef]
- Linder, N.; Konsti, J.; Turkki, R.; Rahtu, E.; Lundin, M.; Nordling, S.; Haglund, C.; Ahonen, T.; Pietikäinen, M.; Lundin, J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn. Pathol. 2012, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Bradish, J.R.; Richey, J.D.; Post, K.M.; Meehan, K.; Sen, J.D.; Malek, A.J.; Katona, T.M.; Warren, S.; Logan, T.F.; Fecher, L.A.; et al. Discordancy in BRAF mutations among primary and metastatic melanoma lesions: Clinical implications for targeted therapy. Mod. Pathol. 2015, 28, 480–486. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK Inhibition versus BRAF Inhibition Alone in Melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossman, P.; Zabka, T.S.; Ruple, A.; Tuerck, D.; Ramos-Vara, J.A.; Liu, L.; Mohallem, R.; Merchant, M.; Franco, J.; Fulkerson, C.M.; et al. Phase I/II trial of vemurafenib in dogs with naturally occurring, BRAF-mutated urothelial carcinoma. Mol. Cancer Ther. 2021, 20, 2177–2188. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, M.; Tambo, N.; Maezawa, M.; Tomihari, M.; Watanabe, K.I.; Inokuma, H.; Miyahara, K. Quantitative analysis of the BRAF V595E mutation in plasma cell-free DNA from dogs with urothelial carcinoma. PLoS ONE 2020, 15, e0232365. [Google Scholar] [CrossRef] [PubMed]
- Aupperle-Lellbach, H.; Grassinger, J.; Hohloch, C.; Kehl, A.; Pantke, P. Diagnostische Aussagekraft der BRAF-Mutation V595E in Urinproben, Ausstrichen und Bioptaten beim kaninen Übergangszellkarzinom. Tierärztliche Prax. Ausg. K Kleintiere Heimtiere 2018, 46, 289–295. [Google Scholar] [CrossRef]
- Wu, S.; Hong, G.; Xu, A.; Zeng, H.; Chen, X.; Wang, Y.; Luo, Y.; Wu, P.; Liu, C.; Jiang, N.; et al. Artificial intelligence-based model for lymph node metastases detection on whole slide images in bladder cancer: A retrospective, multicentre, diagnostic study. Lancet Oncol. 2023, 24, 360–370. [Google Scholar] [CrossRef]
- Fell, C.; Mohammadi, M.; Morrison, D.; Arandjelović, O.; Syed, S.; Konanahalli, P.; Bell, S.; Bryson, G.; Harrison, D.J.; Harris-Birtill, D. Detection of malignancy in whole slide images of endometrial cancer biopsies using artificial intelligence. PLoS ONE 2023, 18, e0282577. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artifi Cial intelligence in cancer research and precision medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Borhani, S.; Borhani, R.; Kajdacsy-Balla, A. Artificial intelligence: A promising frontier in bladder cancer diagnosis and outcome prediction. Crit. Rev. Oncol. Hematol. 2022, 171, 103601. [Google Scholar] [CrossRef] [PubMed]
- Lebret, T.; Pignot, G.; Colombel, M.; Guy, L.; Rebillard, X.; Savareux, L.; Roumigue, M.; Nivet, S.; Coutade Saidi, M.; Piaton, E.; et al. Artificial intelligence to improve cytology performances in bladder carcinoma detection: Results of the VisioCyt test. BJU Int. 2022, 129, 356–363. [Google Scholar] [CrossRef]




| Training Set | Validation Set | |||||
|---|---|---|---|---|---|---|
| No. of slides | 81 | 96 | ||||
| PCR | positive | negative | positive | Negative | ||
| 37 | 44 | 69 | 27 | |||
| Mean ROI | 256 ± 135 mm2 (entire tissue section) | 14 ± 3 mm2 (epithelium only) | ||||
| Quality | high | standard | poor | high | standard | poor |
| 80 | 1 | 0 | 34 | 19 | 43 | |
| PCR positive | 37 | 0 | 0 | 27 | 15 | 27 |
| PCR negative | 43 | 1 | 0 | 7 | 4 | 16 |
| Differentiation | conv. | non-conv. | conv. | non-conv. | ||
| 71 | 10 | 50 | 46 | |||
| PCR positive | 36 | 1 | 40 | 29 | ||
| PCR negative | 35 | 9 | 10 | 17 | ||
| Invasion | present | absent | NA | present | Absent | NA |
| 44 | 36 | 1 | 34 | 23 | 39 | |
| PCR positive | 22 | 15 | 0 | 23 | 19 | 27 |
| PCR negative | 22 | 21 | 1 | 11 | 4 | 12 |
| Inflammation | present | absent | NA | present | Absent | NA |
| 32 | 49 | 0 | 39 | 56 | 1 | |
| PCR positive | 15 | 22 | 0 | 28 | 40 | 1 |
| PCR negative | 17 | 27 | 0 | 11 | 16 | 0 |
| AI Prediction | PCR Result: BRAF Positive | PCR Result: BRAF Negative | Total No. Cases |
|---|---|---|---|
| BRAF positive | 40 | 10 | 50 |
| BRAF negative | 29 | 17 | 46 |
| Total no. cases | 69 | 27 | 96 |
| AI Prediction | No. Cases | |||
|---|---|---|---|---|
| Quality | High | Stand | Poor | |
| BRAF positive | 28 (24) | 8 (7) | 14 (9) | |
| BRAF negative | 6 (3) | 11 (3) | 29 (11) | |
| SE [%]: 89 | SE [%]: 47 | SE [%]: 33 | ||
| SP [%]: 43 | SP [%]: 75 | SP [%]: 69 | ||
| Urothelial differentiation | Present | Absent | ||
| BRAF positive | 42 (34) | 8 (6) | ||
| BRAF negative | 8 (2) | 38 (15) | ||
| SE [%]: 85 | SE [%]: 21 | |||
| SP [%]: 20 | SP [%]: 88 | |||
| Invasive tumour front | Present | Absent | ||
| BRAF positive | 10 (8) | 19 (16) | ||
| BRAF negative | 24 (9) | 4 (1) | ||
| SE [%]: 35 | SE [%]:84 | |||
| SP [%]: 82 | SP [%]: 25 | |||
| Inflammation | Present | Absent | ||
| BRAF positive | 11 (8) | 39 (32) | ||
| BRAF negative | 28 (8) | 17 (9) | ||
| SE [%]: 29 | SE [%]: 80 | |||
| SP [%]: 73 | SP [%]: 56 | |||
| Feature | All Samples | Standard and High-Quality Samples Only | High-Quality Samples Only |
|---|---|---|---|
| Urothelial differentiation | <0.01 | >0.1 | >0.9 |
| Inflammation | <0.01 | <0.03 | >0.7 |
| Slide quality | <0.02 | NA | NA |
| Sample size (ROI) | <0.02 | >0.08 | >0.3 |
| Invasive growth | >0.08 | >0.3 | >0.2 |
| Mutation status (PCR) | >0.6 | >0.1 | <0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Küchler, L.; Posthaus, C.; Jäger, K.; Guscetti, F.; van der Weyden, L.; von Bomhard, W.; Schmidt, J.M.; Farra, D.; Aupperle-Lellbach, H.; Kehl, A.; et al. Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas. Animals 2023, 13, 2404. https://doi.org/10.3390/ani13152404
Küchler L, Posthaus C, Jäger K, Guscetti F, van der Weyden L, von Bomhard W, Schmidt JM, Farra D, Aupperle-Lellbach H, Kehl A, et al. Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas. Animals. 2023; 13(15):2404. https://doi.org/10.3390/ani13152404
Chicago/Turabian StyleKüchler, Leonore, Caroline Posthaus, Kathrin Jäger, Franco Guscetti, Louise van der Weyden, Wolf von Bomhard, Jarno M. Schmidt, Dima Farra, Heike Aupperle-Lellbach, Alexandra Kehl, and et al. 2023. "Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas" Animals 13, no. 15: 2404. https://doi.org/10.3390/ani13152404
APA StyleKüchler, L., Posthaus, C., Jäger, K., Guscetti, F., van der Weyden, L., von Bomhard, W., Schmidt, J. M., Farra, D., Aupperle-Lellbach, H., Kehl, A., Rottenberg, S., & de Brot, S. (2023). Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas. Animals, 13(15), 2404. https://doi.org/10.3390/ani13152404







