Molecular Epidemiology of Pathogenic Leptospira spp. Infecting Dogs in Latin America
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Genetic Data
2.2. Phylogenetic Analyses
3. Results
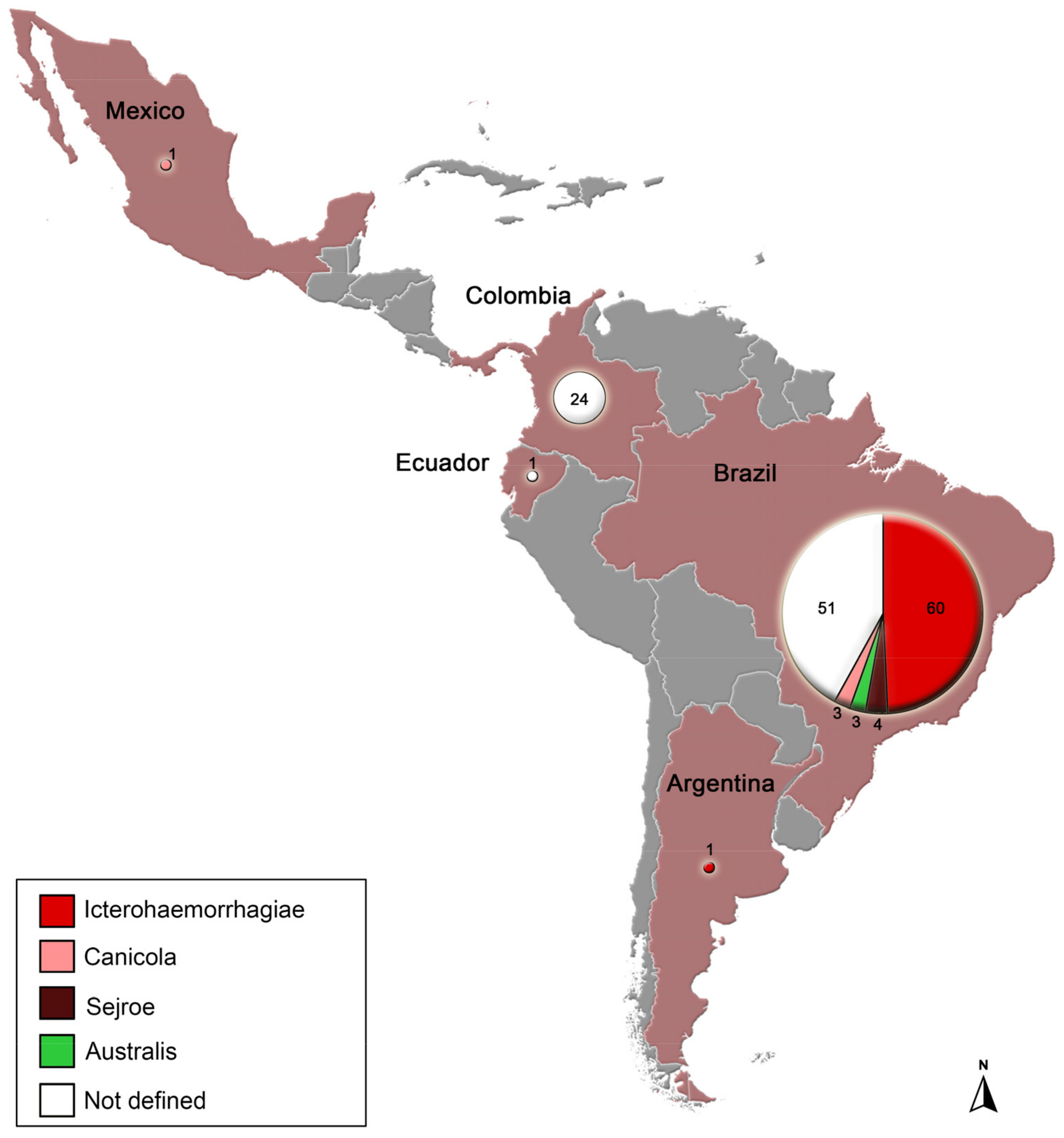
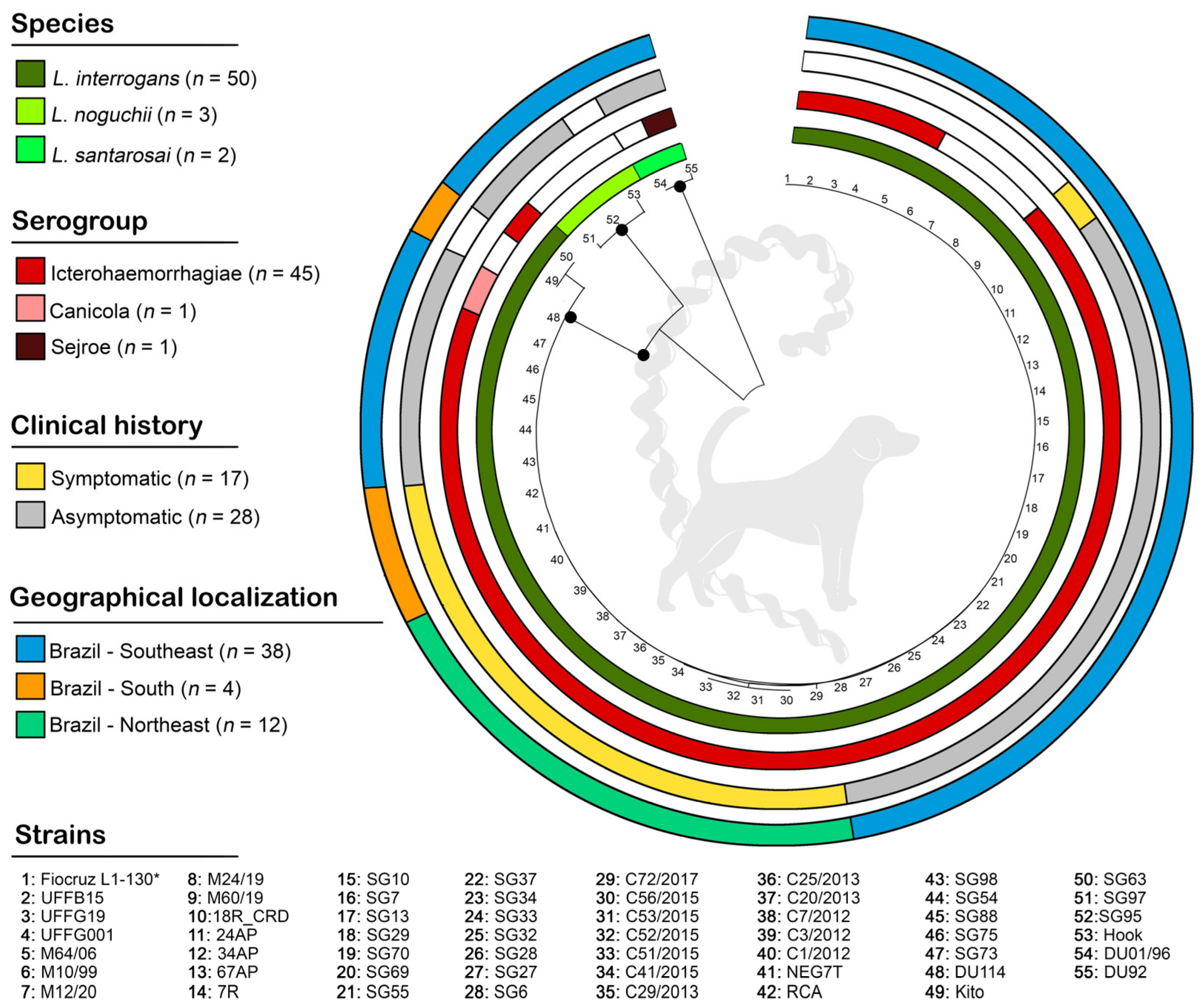
3.1. Description of Genetic Data Obtained
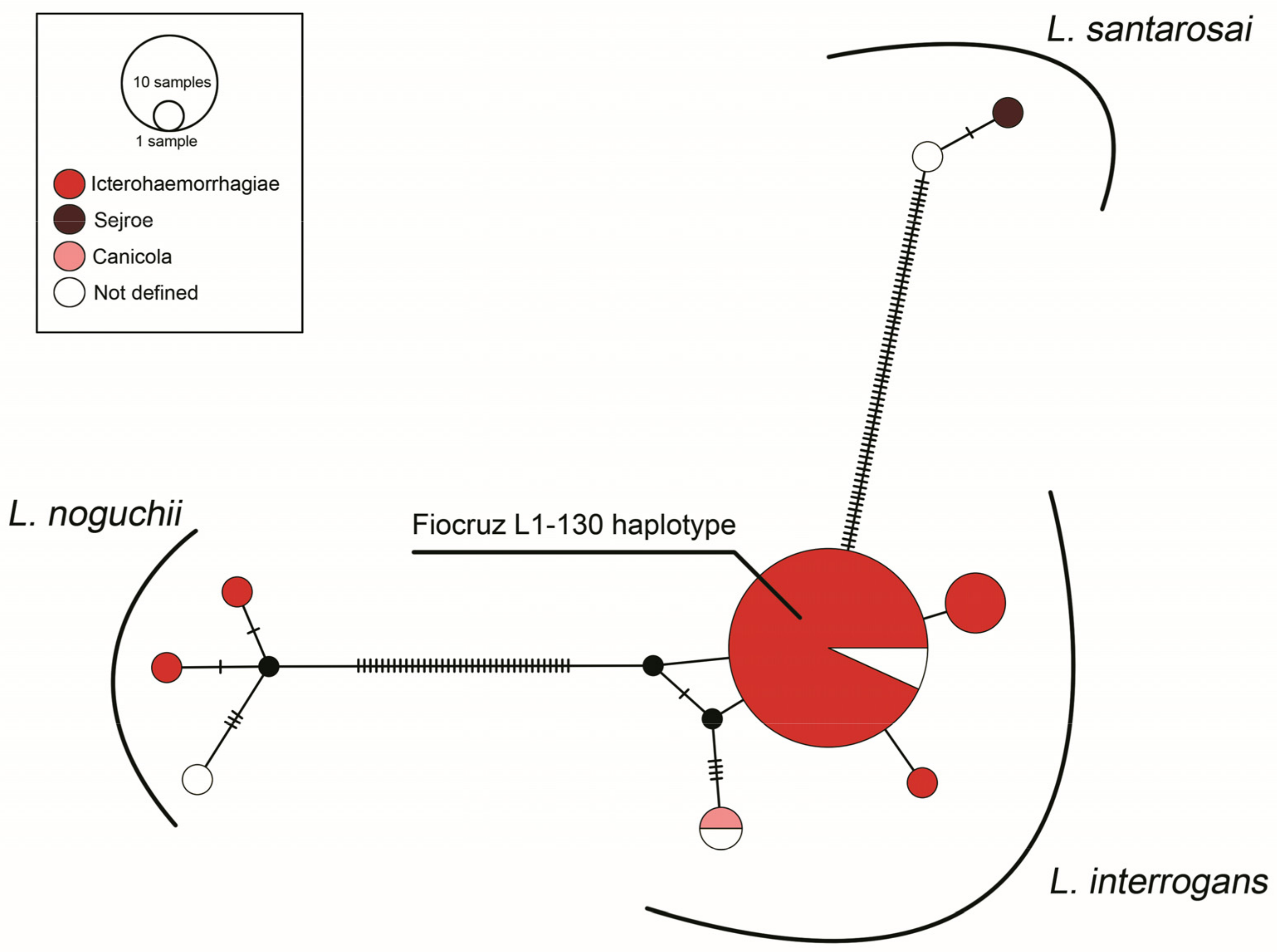
3.2. Molecular Epidemiology Based on Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed]
- Cameron, C.E. Leptospiral Structure, Physiology, and Metabolism. Curr. Top. Microbiol. Immunol. 2015, 387, 21–41. [Google Scholar] [CrossRef]
- Nakamura, S. Motility of the Zoonotic Spirochete Leptospira: Insight into Association with Pathogenicity. Int. J. Mol. Sci. 2022, 23, 1859. [Google Scholar] [CrossRef]
- Samrot, A.V.; Sean, T.C.; Bhavya, K.S.; Sahithya, C.S.; Chan-Drasekaran, S.; Palanisamy, R.; Robinson, E.R.; Subbiah, S.K.; Mok, P.L. Leptospiral Infection, Pathogenesis and Its Diagnosis—A Review. Pathogens 2021, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Systematics of Leptospiraceae. Curr. Top. Microbiol. Immunol. 2015, 387, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the Taxonomy and Evolution of Pathogenicity of the Genus Leptospira through the Prism of Genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Korba, A.A.; Lounici, H.; Kainiu, M.; Vincent, A.T.; Mariet, J.-F.; Veyrier, F.J.; Goarant, C.; Picardeau, M. Leptospira ainlahdjerensis Sp. Nov., Leptospira ainazelensis Sp. Nov., Leptospira abararensis Sp. Nov. and Leptospira chreensis Sp. Nov., Four New Species Isolated from Water Sources in Algeria. Int. J. Syst. Evol. Microbiol. 2021, 71, 005148. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, H.; Li, X.; Cao, Z.; Zhou, H.; Zeng, L.; Xu, J.; Xu, Y.; Chang, Y.-F.; Guo, X.; et al. Molecular Typing of Pathogenic Leptospira Serogroup Icterohaemorrhagiae Strains Circulating in China during the Past 50 Years. PLoS Negl. Trop. Dis. 2015, 9, e0003762. [Google Scholar] [CrossRef]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The Dawn of the Molecular Genetics Era for an Emerging Zoonotic Pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef]
- Ellis, W.A. Animal Leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99–137. [Google Scholar] [CrossRef]
- Santos, C.M.; Dias, G.C.D.R.S.; Saldanha, A.V.P.; Esteves, S.B.; Cortez, A.; Guedes, I.B.; Heinemann, M.B.; Gonçales, A.P.; Miotto, B.A. Molecular and Serological Characterization of Pathogenic Leptospira Spp. Isolated from Symptomatic Dogs in a Highly Endemic Area, Brazil. BMC Vet. Res. 2021, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Haake, D.A.; Levett, P.N. Leptospirosis in Humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [CrossRef]
- Santos, N.J.; Sousa, E.; Reis, M.G.; Ko, A.I.; Costa, F. Rat Infestation Associated with Environmental Deficiencies in an Urban Slum Community with High Risk of Leptospirosis Transmission. Cad. Saúde Pública 2017, 33, e00132115. [Google Scholar] [CrossRef]
- Adler, B.; de la Peña Moctezuma, A. Leptospira and Leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Reagan, K.L.; Sykes, J.E. Diagnosis of Canine Leptospirosis. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Putz, E.J.; Nally, J.E. Investigating the Immunological and Biological Equilibrium of Reservoir Hosts and Pathogenic Leptospira: Balancing the Solution to an Acute Problem? Front. Microbiol. 2020, 11, 2005. [Google Scholar] [CrossRef]
- Adler, B.; Lo, M.; Seemann, T.; Murray, G.L. Pathogenesis of Leptospirosis: The Influence of Genomics. Vet. Microbiol. 2011, 153, 73–81. [Google Scholar] [CrossRef]
- Schuller, S.; Francey, T.; Hartmann, K.; Hugonnard, M.; Kohn, B.; Nally, J.E.; Sykes, J. European Consensus Statement on Leptospirosis in Dogs and Cats. J. Small Anim. Pract. 2015, 56, 159–179. [Google Scholar] [CrossRef]
- Klarenbeek, A.; Schuffner, W.A.P. Appearance in Holland of Leptospira Differing from Weil Strain. Ned. Tijdschr. Geneeskd. 1933, 77, 4271–4276. [Google Scholar]
- Azócar-Aedo, L.; Monti, G. Meta-Analyses of Factors Associated with Leptospirosis in Domestic Dogs. Zoonoses Public Health 2016, 63, 328–336. [Google Scholar] [CrossRef]
- Pinto, P.S.; Libonati, H.; Lilenbaum, W. A Systematic Review of Leptospirosis on Dogs, Pigs, and Horses in Latin America. Trop. Anim. Health Prod. 2017, 49, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna da Costa, R.; Di Azevedo, M.I.N.; Dos Santos Baptista Borges, A.L.; Carvalho-Costa, F.A.; Martins, G.; Lilenbaum, W. Persistent High Leptospiral Shedding by Asymptomatic Dogs in Endemic Areas Triggers a Serious Public Health Concern. Animals 2021, 11, 937. [Google Scholar] [CrossRef]
- Major, A.; Schweighauser, A.; Francey, T. Increasing Incidence of Canine Leptospirosis in Switzerland. Int. J. Environ. Res. Public Health 2014, 11, 7242–7260. [Google Scholar] [CrossRef] [PubMed]
- Sykes, J.E. Chapter 42: Leptospirosis. In Infectious Diseases of the Dog and Cat; Saunders Elsevier: St. Louis, MO, USA, 2012; pp. 431–447. [Google Scholar]
- Raj, J.; Campbell, R.; Tappin, S. Clinical Findings in Dogs Diagnosed with Leptospirosis in England. Vet. Rec. 2021, 189, e452. [Google Scholar] [CrossRef]
- Lippi, I.; Puccinelli, C.; Perondi, F.; Ceccherini, G.; Pierini, A.; Marchetti, V.; Citi, S. Predictors of Fatal Pulmonary Haemorrhage in Dogs Affected by Leptospirosis Approaching Haemodialysis. Vet. Sci. 2021, 8, 25. [Google Scholar] [CrossRef]
- Greenlee, J.J.; Bolin, C.A.; Alt, D.P.; Cheville, N.F.; Andreasen, C.B. Clinical and Pathologic Comparison of Acute Leptospirosis in Dogs Caused by Two Strains of Leptospira kirschneri Serovar Grippotyphosa. Am. J. Vet. Res. 2004, 65, 1100–1107. [Google Scholar] [CrossRef]
- Greenlee, J.J.; Alt, D.P.; Bolin, C.A.; Zuerner, R.L.; Andreasen, C.B. Experimental Canine Leptospirosis Caused by Leptospira interrogans Serovars Pomona and Bratislava. Am. J. Vet. Res. 2005, 66, 1816–1822. [Google Scholar] [CrossRef]
- Esteves, S.B.; Santos, C.M.; Silva, B.C.S.; Salgado, F.F.; Guilloux, A.G.A.; Cortez, A.; Lucco, R.C.; Miotto, B.A. Time for Change? A Systematic Review with Meta-Analysis of Leptospires Infecting Dogs to Assess Vaccine Compatibility in Brazil. Prev. Vet. Med. 2023, 213, 105869. [Google Scholar] [CrossRef]
- Di Azevedo, M.I.N.; Santanna, R.; Carvalho-Costa, F.A.; Lilenbaum, W. The Same Strain Leading to Different Clinical Outcomes: The Enigma behind the Canine Leptospirosis. Microb. Pathog. 2022, 165, 105500. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.A.; Guilloux, A.G.A.; Tozzi, B.F.; Moreno, L.Z.; da Hora, A.S.; Dias, R.A.; Heinemann, M.B.; Moreno, A.M.; de Souza Filho, A.F.; Lilenbaum, W.; et al. Prospective Study of Canine Leptospirosis in Shelter and Stray Dog Populations: Identification of Chronic Carriers and Different Leptospira Species Infecting Dogs. PloS ONE 2018, 13, e0200384. [Google Scholar] [CrossRef]
- do Couto, A.C.; Gravinatti, M.L.; Pellizzaro, M.; Kmetiuk, L.B.; Yamakawa, A.C.; da Silva, E.C.; Felipetto, L.G.; Langoni, H.; de Souza Leandro, A.; de Santi, C.E.; et al. One Health Approach on Serosurvey of Anti-Leptospira Spp. in Homeless Persons and Their Dogs in South Brazil. One Health 2022, 15, 100421. [Google Scholar] [CrossRef] [PubMed]
- Pilau, N.N.; Lubar, A.A.; Daneji, A.I.; Mera, U.M.; Magaji, A.A.; Abiayi, E.A.; Chaiboonma, K.L.; Busayo, E.I.; Vinetz, J.M.; Matthias, M.A. Serological and Molecular Epidemiology of Leptospirosis and the Role of Dogs as Sentinel for Human Infection in Nigeria. Heliyon 2022, 8, e09484. [Google Scholar] [CrossRef] [PubMed]
- Flores, B.J.; Pérez-Sánchez, T.; Fuertes, H.; Sheleby-Elías, J.; Múzquiz, J.L.; Jirón, W.; Duttmann, C.; Halaihel, N. A Cross-Sectional Epidemiological Study of Domestic Animals Related to Human Leptospirosis Cases in Nicaragua. Acta Trop. 2017, 170, 79–84. [Google Scholar] [CrossRef]
- Cárdenas, N.C.; Infante, G.P.; Pacheco, D.A.R.; Diaz, J.P.D.; Wagner, D.C.M.; Dias, R.A.; Neto, J.S.F.; Amaku, M.; Vargas-Pinto, P.; Polo, L.; et al. Seroprevalence of Leptospira Spp. Infection and Its Risk Factors among Domestic Dogs in Bogotá, Colombia. Vet. Anim. Sci. 2018, 6, 64–68. [Google Scholar] [CrossRef]
- Azócar-Aedo, L.; Monti, G. Seroprevalence of Pathogenic Leptospira Spp. in Domestic Dogs from Southern Chile and Risk Factors Associated with Different Environments. Prev. Vet. Med. 2022, 206, 105707. [Google Scholar] [CrossRef] [PubMed]
- Azócar-Aedo, L. Basic Aspects and Epidemiological Studies on Leptospirosis Carried Out in Animals in Chile: A Bibliographic Review. Trop. Med. Infect. Dis. 2023, 8, 97. [Google Scholar] [CrossRef]
- Grune Loffler, S.; Passaro, D.; Samartino, L.; Soncini, A.; Romero, G.; Brihuega, B. Genotypes of Leptospira Spp. Strains Isolated from Dogs in Buenos Aires, Argentina. Rev. Argent. Microbiol. 2014, 46, 201–204. [Google Scholar] [CrossRef]
- Di Azevedo, M.I.N.; Lilenbaum, W. An Overview on the Molecular Diagnosis of Animal Leptospirosis. Lett. Appl. Microbiol. 2021, 72, 496–508. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Paradis, E. Analysis of Haplotype Networks: The Randomized Minimum Spanning Tree Method. Methods Ecol. Evol. 2018, 9, 1308–1317. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Monte Carlo Strategies for Selecting Parameter Values in Simulation Experiments. Syst. Biol. 2015, 64, 741–751. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Guernier, V.; Richard, V.; Nhan, T.; Rouault, E.; Tessier, A.; Musso, D. Leptospira Diversity in Animals and Humans in Tahiti, French Polynesia. PLoS Negl. Trop. Dis. 2017, 11, e0005676. [Google Scholar] [CrossRef] [PubMed]
- López-Osorio, S.; Molano, D.A.; López-Arias, A.; Rodríguez-Osorio, N.; Zambrano, C.; Chaparro-Gutiérrez, J.J. Seroprevalence and Molecular Characterization of Leptospira Spp. in Rats Captured near Pig Farms in Colombia. Int. J. Environ. Res. Public Health 2022, 19, 11539. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.F.; Cerqueira, G.M.; Seyffert, N.; Seixas, F.K.; Hartwig, D.D.; Athanazio, D.A.; Pinto, L.S.; Queiroz, A.; Ko, A.I.; Brod, C.S.; et al. Leptospira noguchii and Human and Animal Leptospirosis, Southern Brazil. Emerg. Infect. Dis. 2009, 15, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Miotto, B.A.; Moreno, L.Z.; Guilloux, A.G.A.; de Sousa, G.O.; Loureiro, A.P.; Moreno, A.M.; Lilenbaum, W.; Vasconcellos, S.A.; Heinemann, M.B.; Hagiwara, M.K. Molecular and Serological Characterization of the First Leptospira santarosai Strain Isolated from a Dog. Acta Trop. 2016, 162, 1–4. [Google Scholar] [CrossRef]
- Perez-Garcia, J.; Monroy, F.P.; Agudelo-Florez, P. Canine Leptospirosis in a Northwestern Region of Colombia: Serological, Molecular and Epidemiological Factors. Pathogens 2022, 11, 1040. [Google Scholar] [CrossRef]
- Zarantonelli, L.; Suanes, A.; Meny, P.; Buroni, F.; Nieves, C.; Salaberry, X.; Briano, C.; Ashfield, N.; Da Silva Silveira, C.; Dutra, F.; et al. Isolation of Pathogenic Leptospira Strains from Naturally Infected Cattle in Uruguay Reveals High Serovar Diversity, and Uncovers a Relevant Risk for Human Leptospirosis. PLoS Negl. Trop. Dis. 2018, 12, e0006694. [Google Scholar] [CrossRef]
- Loureiro, A.P.; Jaeger, L.H.; Di Azevedo, M.I.N.; Miraglia, F.; Moreno, L.Z.; Moreno, A.M.; Pestana, C.P.; Carvalho-Costa, F.A.; Medeiros, M.A.; Lilenbaum, W. Molecular Epidemiology of Leptospira noguchii Reveals Important Insights into a One Health Context. Transbound. Emerg. Dis. 2020, 67, 276–283. [Google Scholar] [CrossRef]
- Aymée, L.; Di Azevedo, M.I.N.; Borges, A.L.D.S.B.; Carvalho-Costa, F.A.; Lilenbaum, W. Leptospira Spp. Strains Associated with Bovine Genital Leptospirosis (BGL). Microb. Pathog. 2022, 173, 105841. [Google Scholar] [CrossRef]
- Lelu, M.; Muñoz-Zanzi, C.; Higgins, B.; Galloway, R. Seroepidemiology of Leptospirosis in Dogs from Rural and Slum Communities of Los Rios Region, Chile. BMC Vet. Res. 2015, 11, 31. [Google Scholar] [CrossRef]
- Benitez, A.D.N.; Monica, T.C.; Miura, A.C.; Romanelli, M.S.; Giordano, L.G.P.; Freire, R.L.; Mitsuka-Breganó, R.; Martins, C.M.; Biondo, A.W.; Serrano, I.M.; et al. Spatial and Simultaneous Seroprevalence of Anti-Leptospira Antibodies in Owners and Their Domiciled Dogs in a Major City of Southern Brazil. Front. Vet. Sci. 2020, 7, 580400. [Google Scholar] [CrossRef]
- Barragan, V.A.; Mejia, M.E.; Trávez, A.; Zapata, S.; Hartskeerl, R.A.; Haake, D.A.; Trueba, G.A. Interactions of Leptospira with Environmental Bacteria from Surface Water. Curr. Microbiol. 2011, 62, 1802–1806. [Google Scholar] [CrossRef] [PubMed]
- Torres, F.D.; Borges, A.L.D.S.B.; Kolesnikovas, C.; Domit, C.; Barbosa, C.B.; Carvalho-Costa, F.A.; Di Azevedo, M.I.N.; Lilenbaum, W. Pinnipeds Carriers of Pathogenic Leptospira: New Data Based on Molecular Characterization. Res. Vet. Sci. 2023, 155, 62–68. [Google Scholar] [CrossRef]
- Caimi, K.; Ruybal, P. Leptospira Spp., a Genus in the Stage of Diversity and Genomic Data Expansion. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 81, 104241. [Google Scholar] [CrossRef]
- Kumaran, S.K.; Bakar, M.F.A.; Mohd-Padil, H.; Mat-Sharani, S.; Sakinah, S.; Poorani, K.; Alsaeedy, H.; Peli, A.; Wei, T.S.; Ling, M.P.; et al. 3D Modelling of the Pathogenic Leptospira Protein LipL32: A Bioinformatics Approach. Acta Trop. 2017, 176, 433–439. [Google Scholar] [CrossRef]
- Jaeger, L.H.; Pestana, C.P.; Carvalho-Costa, F.A.; Medeiros, M.A.; Lilenbaum, W. Characterization of the Clonal Subpopulation Fiocruz L1-130 of Leptospira interrogans in Rats and Dogs from Brazil. J. Med. Microbiol. 2018, 67, 1361–1367. [Google Scholar] [CrossRef]
- Miraglia, F.; Matsuo, M.; Morais, Z.M.; Dellagostin, O.A.; Seixas, F.K.; Freitas, J.C.; Hartskeerl, R.; Moreno, L.Z.; Costa, B.L.; Souza, G.O.; et al. Molecular Characterization, Serotyping, and Antibiotic Susceptibility Profile of Leptospira interrogans Serovar Copenhageni Isolates from Brazil. Diagn. Microbiol. Infect. Dis. 2013, 77, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.M.; Matsuo, M.G.; Bauab, A.R.; Vasconcelos, S.A.; Moraes, Z.M.; Baranton, G.; Saint Girons, I. A Clonal Subpopulation of Leptospira interrogans Sensu Stricto Is the Major Cause of Leptospirosis Outbreaks in Brazil. J. Clin. Microbiol. 2000, 38, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.; Tran, M.-H. One Health: An Effective and Ethical Approach to Leptospirosis Control in Australia. Trop. Med. Infect. Dis. 2022, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- One Health. Available online: https://www.who.int/news-room/questions-and-answers/item/one-health (accessed on 10 June 2023).
- Klaasen, H.L.B.M.; Molkenboer, M.J.C.H.; Vrijenhoek, M.P.; Kaashoek, M.J. Duration of Immunity in Dogs Vaccinated against Leptospirosis with a Bivalent Inactivated Vaccine. Vet. Microbiol. 2003, 95, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna da Costa, R.; Di Azevedo, M.I.N.; Dos Santos Baptista Borges, A.L.; Aymée, L.; Martins, G.; Lilenbaum, W. Effect of Vaccination against Leptospira on Shelter Asymptomatic Dogs Following a Long-Term Study. Animals 2022, 12, 1788. [Google Scholar] [CrossRef]
- Murillo, A.; Cuenca, R.; Serrano, E.; Marga, G.; Ahmed, A.; Cervantes, S.; Caparrós, C.; Vieitez, V.; Ladina, A.; Pastor, J. Leptospira Detection in Cats in Spain by Serology and Molecular Techniques. Int. J. Environ. Res. Public Health 2020, 17, 1600. [Google Scholar] [CrossRef] [PubMed]
- Mota-Rojas, D.; Calderón-Maldonado, N.; Lezama-García, K.; Sepiurka, L.; Maria Garcia, R.C. Abandonment of Dogs in Latin America: Strategies and Ideas. Vet. World 2021, 14, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
| Country | Species | Strain/Sample ID | Clinical History | Serogroup |
|---|---|---|---|---|
| Argentina (n = 1) | L. interrogans (n = 1) | 28 | symptomatic | Icterohaemorrhagiae |
| Brazil (n = 105) | L. interrogans (n = 99) | 18R_CRD, 7R, C1/2012, C20/2013, C25/2013, C29/2013, C3/2012, C41/2015, C51/2015, C52/2015, C53/2015, C56/2015, C7/2012, C72/2017, Dog 10, Dog 16, Dog 2, Dog 9, NEG7, RCA | symptomatic | Icterohaemorrhagiae |
| 24AP, 34AP, 67AP, SG10, SG13, SG27, SG28, SG29, SG3, SG32, SG33, SG34 SG37, SG54, SG55, SG6, SG69, SG7, SG70, SG73, SG75, SG88, SG98 | asymptomatic | |||
| M10/99, M64/06, UFFB15, UFFG001, UFFG19 | unknown | |||
| C80/2017, Dog 6 | symptomatic | Canicola | ||
| DU114 | asymptomatic | Canicola | ||
| Dog 25, Dog 26, Dog 8 | symptomatic | Australis | ||
| Dog 13, Dog 30, Dog 4, Dog 5 | symptomatic | N/D | ||
| 01/014, 01/017, 01/037, 01/070, 01/072, 01/077, 01/078, 01/089, 01/095, 07/070, 13R, 14F, 15F, 16F, 17F, 18F, 21F, 22F, DUZO, H05, H07, H08, H09, H10, H18, H20, H24, H30, H33, H41, H43, H44, Kito, M12/20, M24/19, M60/19, USPIA USPIB, USPIC, USPID, USPIIA | unknown | N/D | ||
| L. noguchii (n = 3) | SG95, SG97 | asymptomatic | N/D | |
| Hook | unknown | N/D | ||
| L. santarosai (n = 3) | DU92, DUPA | asymptomatic | Sejroe | |
| DU01/96 | asymptomatic | N/D | ||
| Colombia (n = 17) | L. interrogans (n = 17) | CfCASol-110-Or, CfCASol-112-Or, CfCASol-135-Or, CfCASol-37-Or, CfCASol-40-As, CfCASol-53-As, CfCASol-59-Or, CfCASol-61-A, CfCASol-72-As, RCA | symptomatic | N/D |
| CfCASol-23-Or, CfCASol-34-O, CfCASol-36-Or, Dog 133, Dog 146, Dog 167, Dog 196 | unknown | N/D | ||
| Ecuador (n = 1) | L.santarosai | Calderón-1 | unknown | N/D |
| Mexico (n = 1) | L. interrogans | LOCaS46 | asymptomatic | Canicola |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Azevedo, M.I.N.; Aymée, L.; Borges, A.L.d.S.B.; Lilenbaum, W. Molecular Epidemiology of Pathogenic Leptospira spp. Infecting Dogs in Latin America. Animals 2023, 13, 2422. https://doi.org/10.3390/ani13152422
Di Azevedo MIN, Aymée L, Borges ALdSB, Lilenbaum W. Molecular Epidemiology of Pathogenic Leptospira spp. Infecting Dogs in Latin America. Animals. 2023; 13(15):2422. https://doi.org/10.3390/ani13152422
Chicago/Turabian StyleDi Azevedo, Maria Isabel Nogueira, Luiza Aymée, Ana Luiza dos Santos Baptista Borges, and Walter Lilenbaum. 2023. "Molecular Epidemiology of Pathogenic Leptospira spp. Infecting Dogs in Latin America" Animals 13, no. 15: 2422. https://doi.org/10.3390/ani13152422
APA StyleDi Azevedo, M. I. N., Aymée, L., Borges, A. L. d. S. B., & Lilenbaum, W. (2023). Molecular Epidemiology of Pathogenic Leptospira spp. Infecting Dogs in Latin America. Animals, 13(15), 2422. https://doi.org/10.3390/ani13152422





